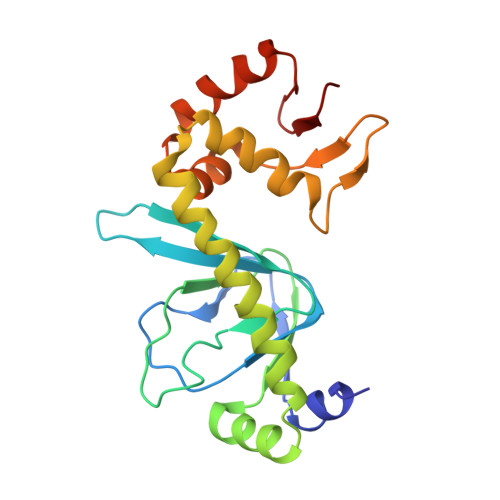
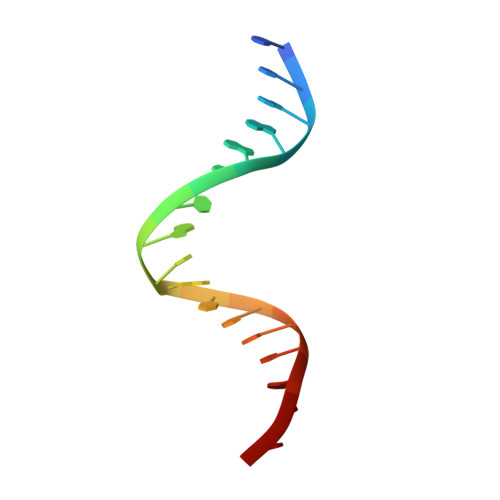
Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex: DNA Binding Specificity Based on Energetics of DNA Kinking
Chen, S., Vojtechovsky, J., Parkinson, G.N., Ebright, R.H., Berman, H.M.(2001) J Mol Biology 314: 63-74
- PubMed: 11724532
- DOI: https://doi.org/10.1006/jmbi.2001.5089
- Primary Citation of Related Structures:
1O3Q, 1O3R, 1O3T - PubMed Abstract:
The catabolite activator protein (CAP) makes no direct contact with the consensus base-pair T:A at position 6 of the DNA half-site 5'-A(1)A(2)A(3)T(4)G(5)T(6)G(7)A(8)T(9)C(10)T(11)-3' but, nevertheless, exhibits strong specificity for T:A at position 6. Binding of CAP results in formation of a sharp DNA kink, with a roll angle of approximately 40 degrees and a twist angle of approximately 20 degrees, between positions 6 and 7 of the DNA half-site. The consensus base-pair T:A at position 6 and the consensus base-pair G:C at position 7 form a T:A/G:C step, which is known to be associated with DNA flexibility. It has been proposed that specificity for T:A at position 6 is a consequence of formation of the DNA kink between positions 6 and 7, and of effects of the T:A(6)/G:C(7) step on the geometry of DNA kinking, or the energetics of DNA kinking. In this work, we determine crystallographic structures of CAP-DNA complexes having the consensus base-pair T:A at position 6 or the non-consensus base-pair C:G at position 6. We show that complexes containing T:A or C:G at position 6 exhibit similar overall DNA bend angles and local geometries of DNA kinking. We infer that indirect readout in this system does not involve differences in the geometry of DNA kinking but, rather, solely differences in the energetics of DNA kinking. We further infer that the main determinant of DNA conformation in this system is protein-DNA interaction, and not DNA sequence.
- Department of Chemistry and The Waksman Institute, Rutgers, the State University of New Jersey, 610 Taylor Road, Piscataway, NJ 08854-8087, USA.
Organizational Affiliation: