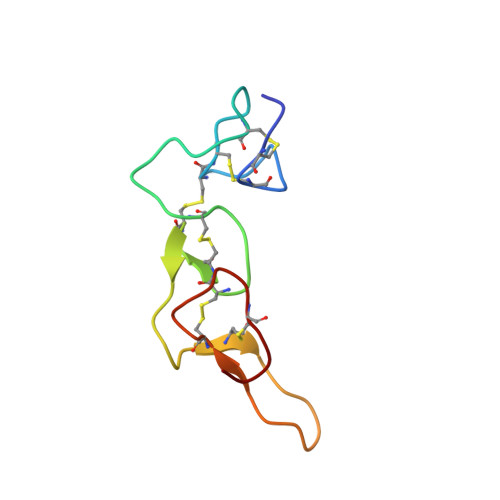
Solution structure of kistrin, a potent platelet aggregation inhibitor and GP IIb-IIIa antagonist.
Adler, M., Lazarus, R.A., Dennis, M.S., Wagner, G.(1991) Science 253: 445-448
- PubMed: 1862345
- DOI: https://doi.org/10.1126/science.1862345
- Primary Citation of Related Structures:
1N4Y - PubMed Abstract:
The structure of kistrin, which is a member of a homologous family of glycoprotein IIb-IIIa (GP IIb-IIIa) antagonists and potent protein inhibitors of platelet aggregation, has been determined by two-dimensional nuclear magnetic resonance (NMR) spectroscopy. The 68-residue protein consists of a series of tightly packed loops held together by six disulfide bonds and has almost no regular secondary structure. Kistrin has an Arg-Gly-Asp (RGD) adhesion site recognition sequence important for binding to GP IIb-IIIa that is located at the apex of a long loop across the surface of the protein.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115.
Organizational Affiliation: