Structural comparison of allogeneic and syngeneic T cell receptor-peptide-major histocompatibility complex complexes: a buried alloreactive mutation subtly alters peptide presentation substantially increasing V(beta) Interactions.
Luz, J.G., Huang, M.D., Garcia, K.C., Rudolph, M.G., Teyton, L., Wilson, I.A.(2002) J Exp Medicine 195: 1175-1186
- PubMed: 11994422
- DOI: https://doi.org/10.1084/jem.20011644
- Primary Citation of Related Structures:
1LEG, 1LEK, 1MWA - PubMed Abstract:
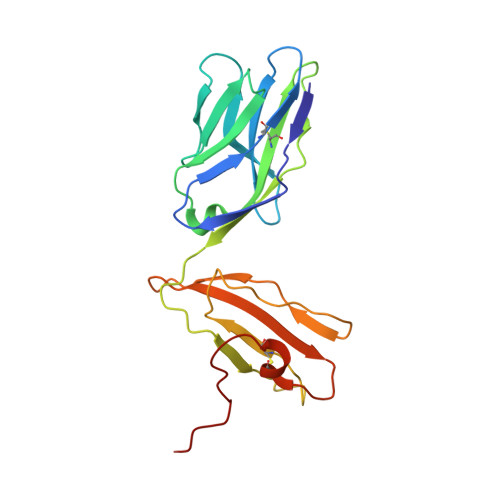
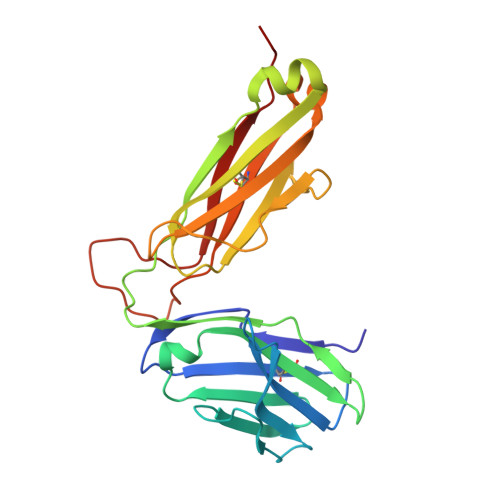
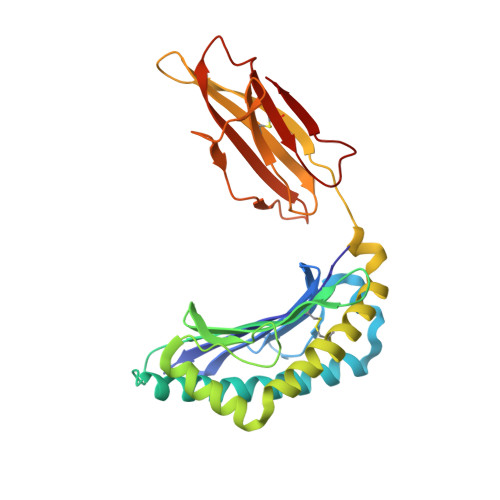
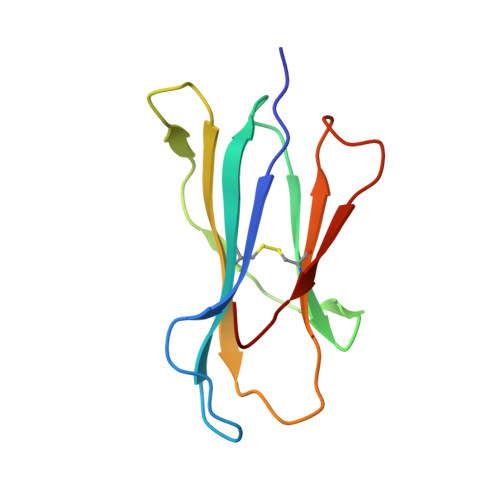
The crystal structures of the 2C/H-2K(bm3)-dEV8 allogeneic complex at 2.4 A and H-2K(bm3)-dEV8 at 2.15 A, when compared with their syngeneic counterparts, elucidate structural changes that induce an alloresponse. The Asp77Ser mutation that imbues H-2K(bm3)-dEV8 with its alloreactive properties is located beneath the peptide and does not directly contact the T cell receptor (TCR). However, the buried mutation induces local rearrangement of the peptide itself to preserve hydrogen bonding interactions between the peptide and the alpha(1) 77 residue. The COOH terminus of the peptide main chain is tugged toward the alpha(1)-helix such that its presentation to the TCR is altered. These changes increase the stability of the allogeneic peptide-major histocompatibility complex (pMHC) complex and increase complementarity in the TCR-pMHC interface, placing greater emphasis on recognition of the pMHC by the TCR beta-chain, evinced by an increase in shape complementarity, buried surface area, and number of TCR-pMHC contacting residues. A nearly fourfold increase in the number of beta-chain-pMHC contacts is accompanied by a concomitant 64% increase in beta-chain-pMHC shape complementarity. Thus, the allogeneic mutation causes the same peptide to be presented differently, temporally and spatially, by the allogeneic and syngeneic MHCs.
- Department of Molecular Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: