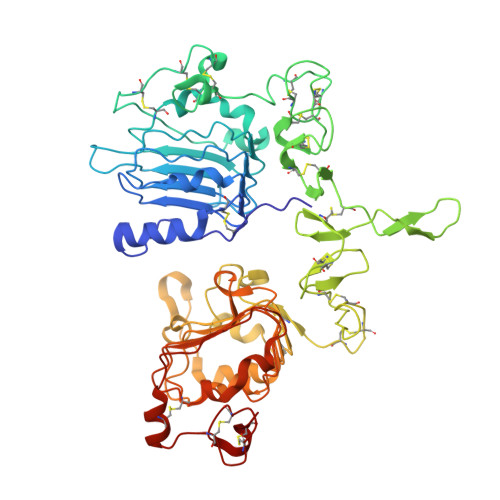
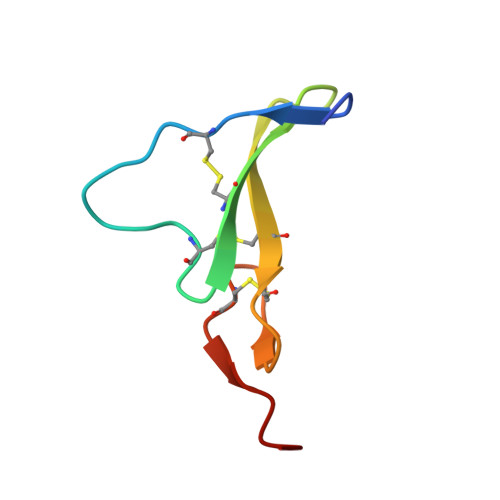
Crystal Structure of a Truncated Epidermal Growth Factor Receptor Extracellular Domain Bound to Transforming Growth Factor alpha
Garrett, T.P.J., McKern, N.M., Lou, M., Elleman, T.C., Adams, T.E., Lovrecz, G.O., Zhu, H.-J., Walker, F., Frenkel, M.J., Hoyne, P.A., Jorissen, R.N., Nice, E.C., Burgess, A.W., Ward, C.W.(2002) Cell 110: 763-773
- PubMed: 12297049
- DOI: https://doi.org/10.1016/s0092-8674(02)00940-6
- Primary Citation of Related Structures:
1MOX - PubMed Abstract:
We report the crystal structure, at 2.5 A resolution, of a truncated human EGFR ectodomain bound to TGFalpha. TGFalpha interacts with both L1 and L2 domains of EGFR, making many main chain contacts with L1 and interacting with L2 via key conserved residues. The results indicate how EGFR family members can bind a family of highly variable ligands. In the 2:2 TGFalpha:sEGFR501 complex, each ligand interacts with only one receptor molecule. There are two types of dimers in the asymmetric unit: a head-to-head dimer involving contacts between the L1 and L2 domains and a back-to-back dimer dominated by interactions between the CR1 domains of each receptor. Based on sequence conservation, buried surface area, and mutagenesis experiments, the back-to-back dimer is favored to be biologically relevant.
- Walter and Eliza Hall Institute of Medical Research, Post Office Royal Melbourne Hospital, Parkville, 3050, Victoria, Australia. tgarrett@wehi.edu.au
Organizational Affiliation: