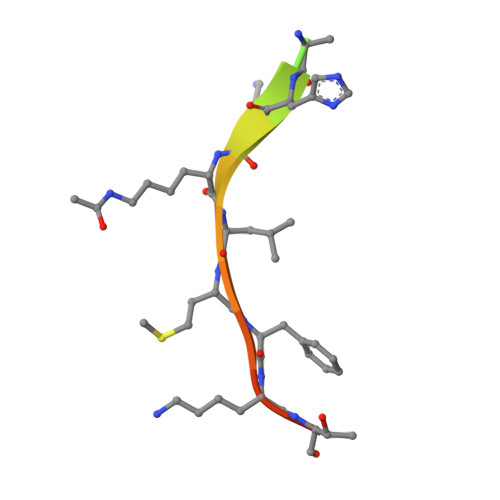
Structure of a Sir2 enzyme bound to an acetylated p53 peptide
Avalos, J.L., Celic, I., Muhammad, S., Cosgrove, M.S., Boeke, J.D., Wolberger, C.(2002) Mol Cell 10: 523-535
- PubMed: 12408821
- DOI: https://doi.org/10.1016/s1097-2765(02)00628-7
- Primary Citation of Related Structures:
1MA3 - PubMed Abstract:
Sir2 proteins are NAD(+)-dependent protein deacetylases that play key roles in transcriptional regulation, DNA repair, and life span regulation. The structure of an archaeal Sir2 enzyme, Sir2-Af2, bound to an acetylated p53 peptide reveals that the substrate binds in a cleft in the enzyme, forming an enzyme-substrate beta sheet with two flanking strands in Sir2-Af2. The acetyl-lysine inserts into a conserved hydrophobic tunnel that contains the active site histidine. Comparison with other structures of Sir2 enzymes suggests that the apoenzyme undergoes a conformational change upon substrate binding. Based on the Sir2-Af2 substrate complex structure, mutations were made in the other A. fulgidus sirtuin, Sir2-Af1, that increased its affinity for the p53 peptide.
- Department of Biophysics and Biophysical Chemistry, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Organizational Affiliation: