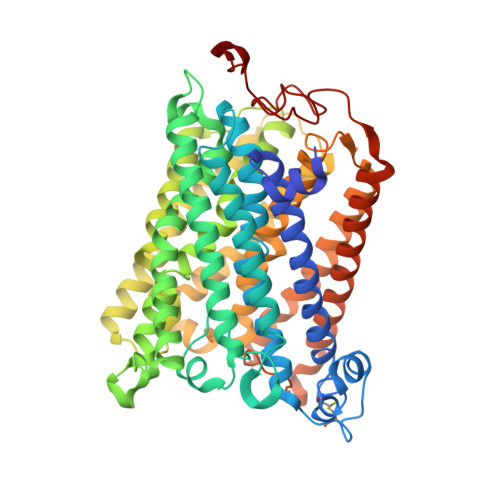
The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides.
Svensson-Ek, M., Abramson, J., Larsson, G., Tornroth, S., Brzezinski, P., Iwata, S.(2002) J Mol Biology 321: 329-339
- PubMed: 12144789
- DOI: https://doi.org/10.1016/s0022-2836(02)00619-8
- Primary Citation of Related Structures:
1M56, 1M57 - PubMed Abstract:
The structure of cytochrome c oxidase from Rhodobacter sphaeroides has been solved at 2.3/2.8A (anisotropic resolution). This high-resolution structure revealed atomic details of a bacterial terminal oxidase including water molecule positions and a potential oxygen pathway, which has not been reported in other oxidase structures. A comparative study of the wild-type and the EQ(I-286) mutant enzyme revealed structural rearrangements around E(I-286) that could be crucial for proton transfer in this enzyme. In the structure of the mutant enzyme, EQ(I-286), which cannot transfer protons during oxygen reduction, the side-chain of Q(I-286) does not have the hydrogen bond to the carbonyl oxygen of M(I-107) that is seen in the wild-type structure. Furthermore, the Q(I-286) mutant has a different arrangement of water molecules and residues in the vicinity of the Q side-chain. These differences between the structures could reflect conformational changes that take place upon deprotonation of E(I-286) during turnover of the wild-type enzyme, which could be part of the proton-pumping machinery of the enzyme.
- Department of Biochemistry, Biomedical Center, Uppsala University, Box 576, Uppsala, Sweden. margareta.e.ek@astrazeneca.com
Organizational Affiliation: