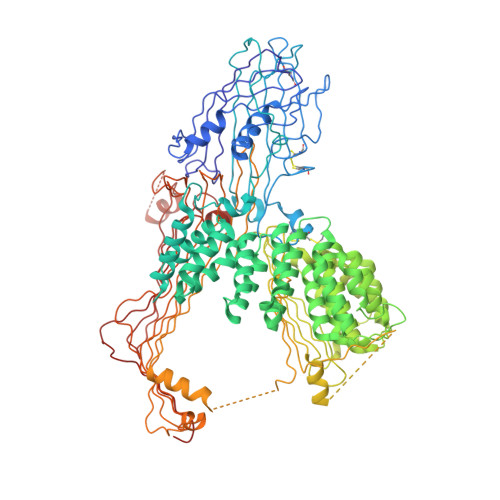
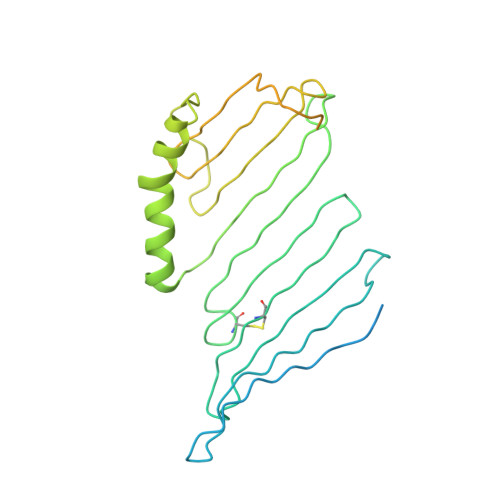
Lipid-protein interactions in lipovitellin.
Thompson, J.R., Banaszak, L.J.(2002) Biochemistry 41: 9398-9409
- PubMed: 12135361
- DOI: https://doi.org/10.1021/bi025674w
- Primary Citation of Related Structures:
1LSH - PubMed Abstract:
The refined molecular structure of lipovitellin is described using synchrotron cryocrystallographic data to 1.9 A resolution. Lipovitellin is the predominant lipoprotein found in the yolk of egg-laying animals and is involved in lipid and metal storage. It is thought to be related in amino acid sequence to segments of apolipoprotein B and the microsomal transfer protein responsible for the assembly of low-density lipoproteins. Lipovitellin contains a heterogeneous mixture of about 16% (w/w) noncovalently bound lipid, mostly phospholipid. Previous X-ray structural studies at ambient temperature described several different protein domains including a large cavity in each subunit of the dimeric protein. The cavity was free of any visible electron density for lipid molecules at room temperature, suggesting that only dynamic interactions exist with the protein. An important result from this crystallographic study at 100 K is the appearance of some bound ordered lipid along the walls of the binding cavity. The precise identification of the lipid type is difficult because of discontinuities in the electron density. Nonetheless, the conformations of 7 phospholipids and 43 segments of hydrocarbon chains greater than 5 atoms in length have been discovered. The conformations of the bound lipid and the interactions between protein and lipid provide insights into the factors governing lipoprotein formation.
- Department of Biochemistry, Molecular Biology, and Biophysics, University of Minnesota, Minneapolis, Minnesota 55455, USA.
Organizational Affiliation: