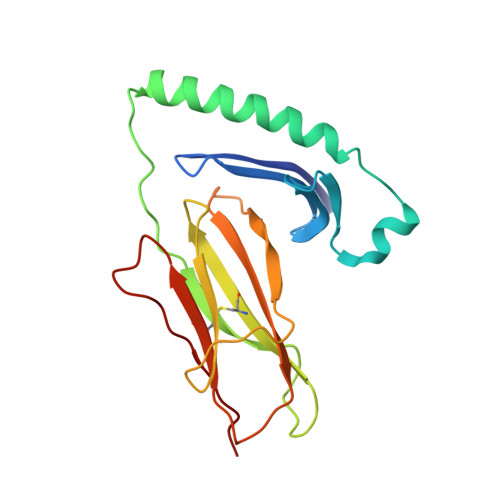
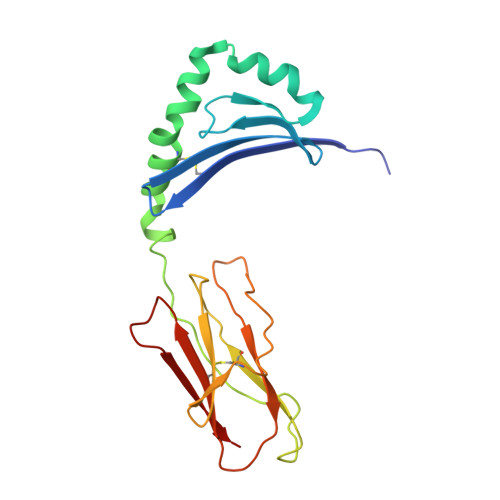
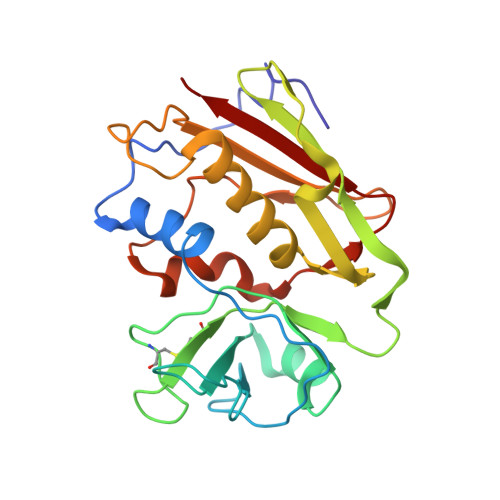
Crystal Structure of a SEA Variant in Complex with MHC Class II Reveals the Ability of SEA to Crosslink MHC Molecules
Petersson, K., Thunnissen, M., Forsberg, G., Walse, B.(2002) Structure 10: 1619-1626
- PubMed: 12467569
- DOI: https://doi.org/10.1016/s0969-2126(02)00895-x
- Primary Citation of Related Structures:
1LO5 - PubMed Abstract:
Although the biological properties of staphylococcal enterotoxin A (SEA) have been well characterized, structural insights into the interaction between SEA and major histocompatibilty complex (MHC) class II have only been obtained by modeling. Here, the crystal structure of the D227A variant of SEA in complex with human MHC class II has been determined by X-ray crystallography. SEA(D227A) exclusively binds with its N-terminal domain to the alpha chain of HLA-DR1. The ability of one SEA molecule to crosslink two MHC molecules was modeled. It shows that this SEA molecule cannot interact with the T cell receptor (TCR) while a second SEA molecule interacts with MHC. Because of its relatively low toxicity, the D227A variant of SEA is used in tumor therapy.
- Molecular Biophysics, Centre for Chemistry and Chemical Engineering, Lund University, P.O. Box 124, S-221 00 Lund, Sweden.
Organizational Affiliation: