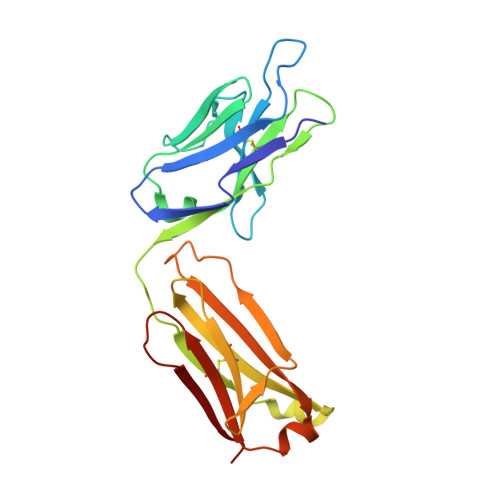
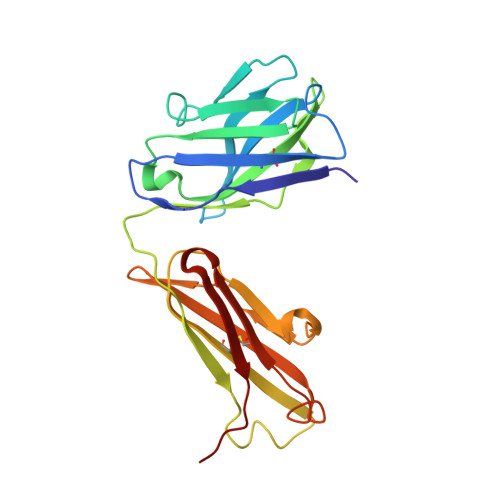
A structural basis for the activity of retro-Diels-Alder catalytic antibodies: evidence for a catalytic aromatic residue.
Hugot, M., Bensel, N., Vogel, M., Reymond, M.T., Stadler, B., Reymond, J.L., Baumann, U.(2002) Proc Natl Acad Sci U S A 99: 9674-9678
- PubMed: 12093912
- DOI: https://doi.org/10.1073/pnas.142286599
- Primary Citation of Related Structures:
1LO0, 1LO2, 1LO3, 1LO4 - PubMed Abstract:
The nitroxyl synthase catalytic antibodies 10F11, 9D9, and 27C5 catalyze the release of nitroxyl from a bicyclic pro-drug by accelerating a retro-Diels-Alder reaction. The Fabs (antigen-binding fragments) of these three catalytic antibodies were cloned and sequenced. Fab 9D9 was crystallized in the apo-form and in complex with one transition state analogue of the reaction. Crystal structures of Fab 10F11 in complex with ligands mimicking substrate, transition state, and product have been determined at resolutions ranging from 1.8 to 2.3 A. Antibodies 9D9 and 10F11 show increased shape complementarity (as quantified by the program sc) to the hapten and to a modeled transition state as compared with substrate and product. The shape complementarity is mediated to a large extent by an aromatic residue (tyrosine or tryptophan) at the bottom of the hydrophobic active pocket, which undergoes pi-stacking interactions with the aromatic rings of the ligands. Another factor contributing to the different reactivity of the regioisomers probably arises because of hydrogen-bonding interactions between the nitroxyl bridge and the backbone amide of PheH101 and possibly a conserved water molecule.
- Department of Chemistry and Biochemistry, University of Berne, Freiestrasse 3, CH-3012 Berne, Switzerland.
Organizational Affiliation: