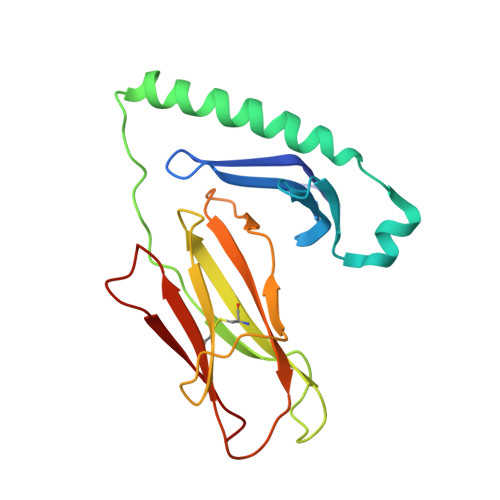
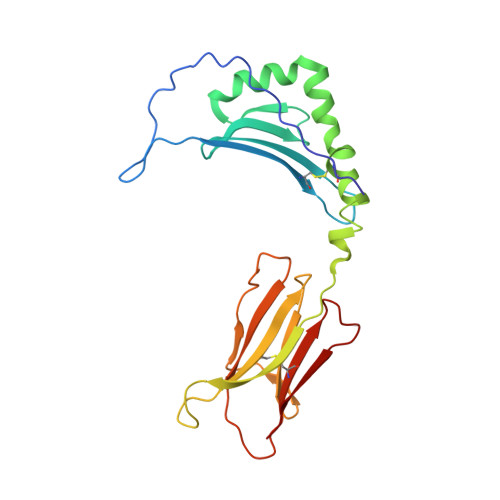
Alternate interactions define the binding of peptides to the MHC molecule IA(b).
Liu, X., Dai, S., Crawford, F., Fruge, R., Marrack, P., Kappler, J.(2002) Proc Natl Acad Sci U S A 99: 8820-8825
- PubMed: 12084926
- DOI: https://doi.org/10.1073/pnas.132272099
- Primary Citation of Related Structures:
1LNU - PubMed Abstract:
We have solved the crystal structure of the MHCII molecule, IA(b), containing an antigenic variant of the major IA(b)-binding peptide derived from the MHCII IEalpha chain. The four MHC pockets at p1, p4, p6, and p9 that usually bind peptide side chains are largely empty because of alanines in the peptide at these positions. The complex is nevertheless very stable, apparently because of unique alternate interactions between the IA(b) and peptide. In particular, there are multiple additional hydrogen bonds between the N-terminal end of the peptide and the IA(b) alpha chain and an extensive hydrogen bond network involving an asparagine at p7 position of the peptide and the IA(b) beta chain. By using knowledge of the shape and size of the traditional side chain binding pockets and the additional possible interactions, an IA(b) peptide-binding motif can be deduced that agrees well with the sequences of known IA(b)-binding peptides.
- Integrated Department of Immunology, Zuckerman Family/Canyon Ranch Crystallography Laboratory, Howard Hughes Medical Institute, National Jewish Medical and Research Center, 1400 Jackson Street, Denver, CO 80206, USA.
Organizational Affiliation: