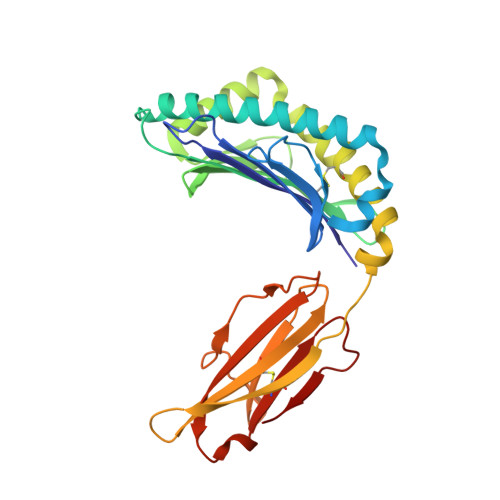
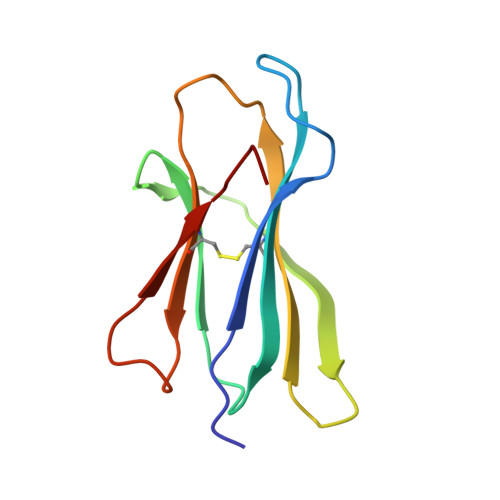
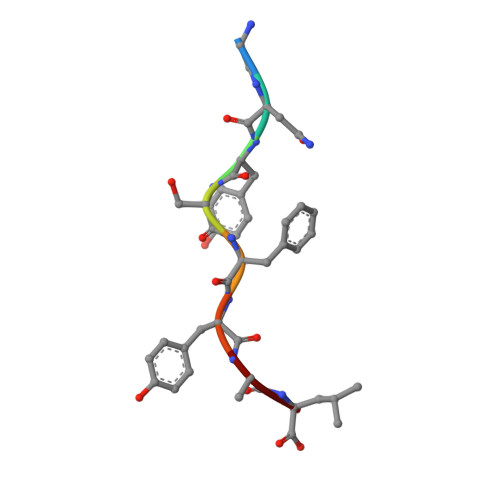
A peptide that antagonizes TCR-mediated reactions with both syngeneic and allogeneic agonists: functional and structural aspects.
Rudolph, M.G., Shen, L.Q., Lamontagne, S.A., Luz, J.G., Delaney, J.R., Ge, Q., Cho, B.K., Palliser, D., McKinley, C.A., Chen, J., Wilson, I.A., Eisen, H.N.(2004) J Immunol 172: 2994-3002
- PubMed: 14978103
- DOI: https://doi.org/10.4049/jimmunol.172.5.2994
- Primary Citation of Related Structures:
1LK2 - PubMed Abstract:
We identify and consider some characteristics of a peptide antagonist for the Ag-specific receptor on 2C cells (the 2C TCR). The peptide, GNYSFYAL (called GNY), binds to H-2K(b), and a very high-resolution crystal structure of the GNY-K(b) complex at 1.35 A is described. Although the GNY peptide does not bind to L(d), the potency of GNY-K(b) as an antagonist is evident from its ability to specifically inhibit 2C TCR-mediated reactions to an allogenic agonist complex (QLSPFPFDL-L(d)), as well as to a syngeneic agonist complex (SIYRYYGL-K(b)). The crystal structure and the activities of alanine-substituted peptide variants point to the properties of the peptide P4 side chain and the conformation of the Tyr-P6 side chain as the structural determinants of GNYSFYAL antagonist activity.
- Center for Cancer Research and Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Markus.Rudolph@bio.uni-goettingen.de
Organizational Affiliation: