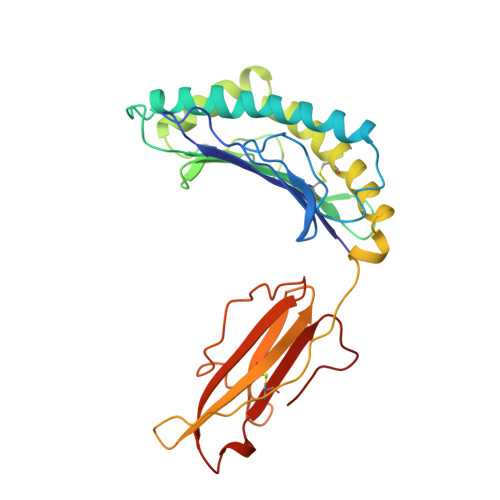
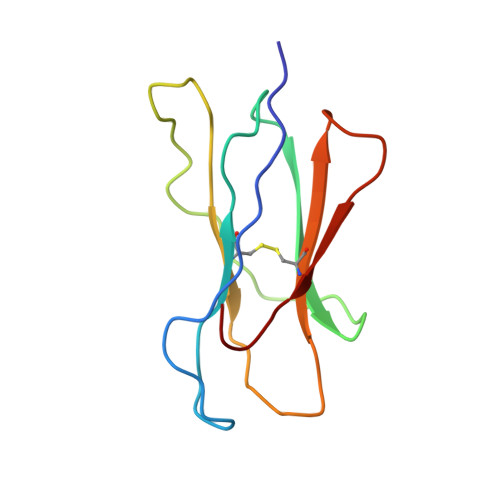
Structural basis of 2C TCR allorecognition of H-2Ld peptide complexes.
Speir, J.A., Garcia, K.C., Brunmark, A., Degano, M., Peterson, P.A., Teyton, L., Wilson, I.A.(1998) Immunity 8: 553-562
- PubMed: 9620676
- DOI: https://doi.org/10.1016/s1074-7613(00)80560-9
- Primary Citation of Related Structures:
1LDP - PubMed Abstract:
MHC class I H-2Ld complexed with peptide QL9 (or p2Ca) is a high-affinity alloantigen for the 2C TCR. We used the crystal structure of H-2Ld with a mixture of bound peptides at 3.1 A to construct a model of the allogeneic 2C-Ld/QL9 complex for comparison with the syngeneic 2C-Kb/dEV8 structure. A prominent ridge on the floor of the Ld peptide-binding groove, not present in Kb, creates a C-terminal bulge in Ld peptides that greatly increases interactions with the 2C beta-chain. Furthermore, weak electrostatic complementarity between Asp77 on the alpha1 helix of Kb and 2C is enhanced in the allogeneic complex by closer proximity of QL9 peptide residue AspP8 to the 2C HV4 loop.
- Department of Molecular Biology, The Scripps Research Institute, La Jolla, California 92037, USA.
Organizational Affiliation: