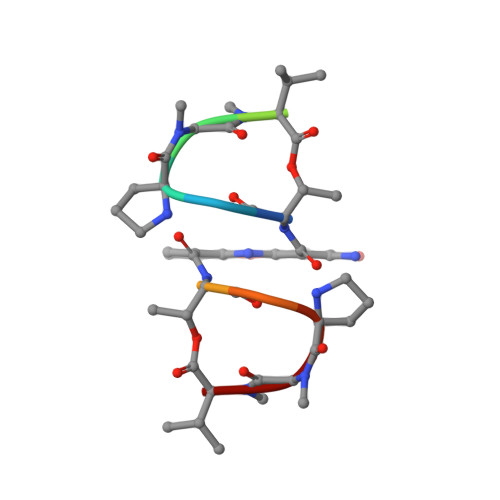
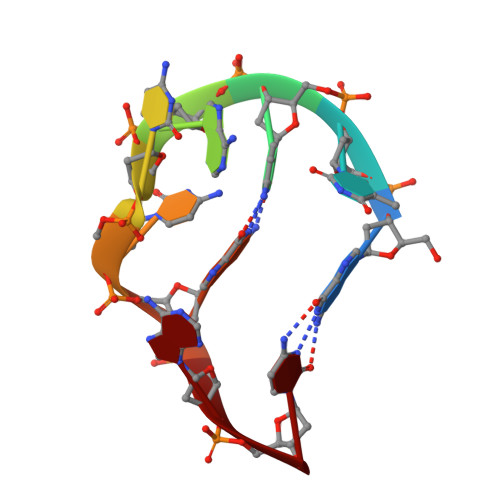
Looped Out and Perpendicular: Deformation of Watson-Crick Base Pair Associated with Actinomycin D Binding.
Chou, S.H., Chin, K.H., Chen, F.M.(2002) Proc Natl Acad Sci U S A 99: 6625
- PubMed: 12011426
- DOI: https://doi.org/10.1073/pnas.102580399
- Primary Citation of Related Structures:
1L1V - PubMed Abstract:
Many anticancer drugs interact directly with DNA to exert their biological functions. To date, all noncovalent, intercalating drugs interact with DNA exclusively by inserting their chromophores into base steps to form elongated and unwound duplex structures without disrupting the flanking base pairs. By using actinomycin D (ActD)-5'-GXC/CYG-5' complexes as examples, we have found a rather unusual interaction mode for the intercalated drug; the central Watson-Crick X/Y base pairs are looped out and displaced by the ActD chromophore. The looped-out bases are not disordered but interact perpendicularly with the base/chromophore and form specific H bonds with DNA. Such a complex structure provides intriguing insights into how ligand interacts with DNA and enlarges the repertoires for sequence-specific DNA recognition.
- Institute of Biochemistry, National Chung-Hsing University, Taichung, Taiwan 40227, Republic of China. shchou@dragon.nchu.edu.tw
Organizational Affiliation: