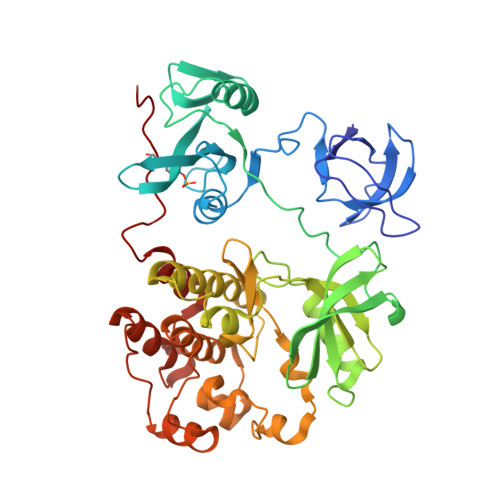
Mutant tyrosine kinases with unnatural nucleotide specificity retain the structure and phospho-acceptor specificity of the wild-type enzyme.
Witucki, L.A., Huang, X., Shah, K., Liu, Y., Kyin, S., Eck, M.J., Shokat, K.M.(2002) Chem Biol 9: 25-33
- PubMed: 11841936
- DOI: https://doi.org/10.1016/s1074-5521(02)00091-1
- Primary Citation of Related Structures:
1KSW - PubMed Abstract:
The direct substrates of one protein kinase in a cell can be identified by mutation of the ATP binding pocket to allow an unnatural ATP analog to be accepted exclusively by the engineered kinase. Here, we present structural and functional assessment of peptide specificity of mutant protein kinases with unnatural ATP analogs. The crystal structure (2.8 A resolution) of c-Src (T338G) with N(6)-(benzyl) ADP bound shows that the creation of a unique nucleotide binding pocket does not alter the phospho-acceptor binding site of the kinase. A panel of optimal peptide substrates of defined sequence, as well as a degenerate peptide library, was utilized to assess the phospho-acceptor specificity of the engineered "traceable" kinases. The specificity profiles for the mutant kinases were found to be identical to those of their wild-type counterparts.
- Department of Cellular and Molecular Pharmacology, Box 0450, University of California, San Francisco 94143-0450, USA.
Organizational Affiliation: