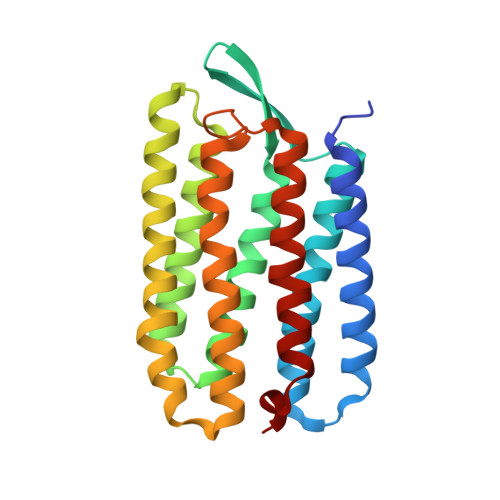
Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure.
Faham, S., Bowie, J.U.(2002) J Mol Biology 316: 1-6
- PubMed: 11829498
- DOI: https://doi.org/10.1006/jmbi.2001.5295
- Primary Citation of Related Structures:
1KME - PubMed Abstract:
Obtaining crystals of membrane proteins that diffract to high resolution remains a major stumbling block in structure determination. Here we present a new method for crystallizing membrane proteins from a bicelle forming lipid/detergent mixture. The method is flexible and simple to use. As a test case, bacteriorhodopsin (bR) from Halobacterium salinarum was crystallized from a bicellar solution, yielding a new bR crystal form. The crystals belong to space group P2(1) with unit cell dimensions of a=45.0 A, b=108.9 A, c=55.9 A, beta=113.58 degrees and a dimeric asymmetric unit. The structure was solved by molecular replacement and refined at 2.0 A resolution. In all previous bR structures the protein is organized as a parallel trimer, but in the crystals grown from bicelles, the individual bR subunits are arranged in an antiparallel fashion.
- Department of Chemistry and Biochemistry, University of California, Los Angeles, CA 90095-1570, USA.
Organizational Affiliation: