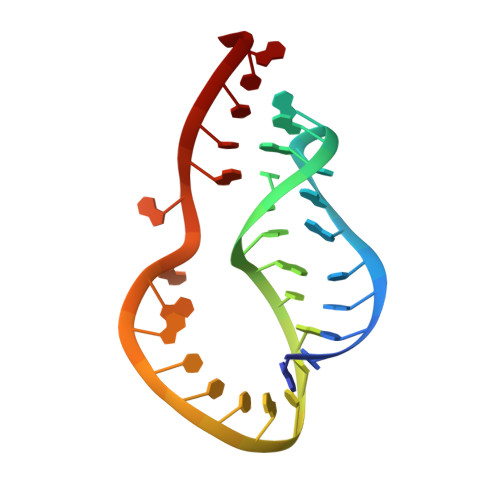
Conformation of a non-frameshifting RNA pseudoknot from mouse mammary tumor virus.
Kang, H., Hines, J.V., Tinoco Jr., I.(1996) J Mol Biology 259: 135-147
- PubMed: 8648641
- DOI: https://doi.org/10.1006/jmbi.1996.0308
- Primary Citation of Related Structures:
1KAJ - PubMed Abstract:
The solution conformation of an RNA pseudoknot, which is a mutant of the pseudoknot required for ribosomal frameshifting in mouse mammary tumor virus, has been determined by NMR. The 32-nucleotide RNA pseudoknot does not promote efficient frameshifting, although its sequence is very similar to the efficient frameshifting pseudoknot whose structure was recently determined by our group. 13C-labeling of the RNA and 13C-edited NMR techniques were used to facilitate spectral assignment. The three-dimensional structure of the RNA pseudoknot was determined by restrained molecular dynamics based on NMR-derived interproton distances and torsion angle constraints. The conformation is very different from that previously determined for the efficient-frameshifting pseudoknot. Two unpaired nucleotides are stacked between stem 1 and stem 2, in contrast to the one unpaired nucleotide at the same junction region as found previously. The two stems of the pseudoknot are not coaxial, they are twisted and bent relative to each other. Loop 2 does not cross the shallow minor groove of stem 1, in contrast to the pseudoknots with one or no intervening nucleotides between the stems. The fact that a specific conformation is required for efficient frameshifting implies a specific interaction of the pseudoknot with the ribosome.
- Department of Chemistry, University of California, Berkeley, USA.
Organizational Affiliation: