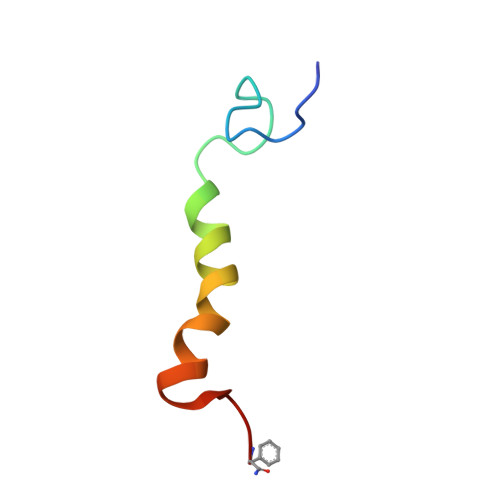
The NMR-derived conformation of neuropeptide F from Moniezia expansa.
Miskolzie, M., Kotovych, G.(2002) J Biomol Struct Dyn 19: 991-998
- PubMed: 12023801
- DOI: https://doi.org/10.1080/07391102.2002.10506802
- Primary Citation of Related Structures:
1K8V - PubMed Abstract:
The solution structure of neuropeptide F (NPF), from the flatworm (platyhelminthes) Moniezia expansa, has been determined by (1)H NMR spectroscopy at 800 MHz in 60%/40% CD(3)OH/H(2)O. NPF is the most abundant neuropeptide in platyhelminthes. The secondary structure of NPF contains an alpha helix from residues Lys(14) to Ile(31), while the N terminus, consisting of residues Pro(-2) to Asn(13), and the C-terminus, consisting of residues Gly(32) to Phe(36), are in a random conformation. The structure was calculated by a simulated annealing protocol, and the conformational data are compared to the porcine neuropeptide Y (NPY), a peptide hormone and neurotransmitter. The exact function of NPF is unknown, but structural similarity with porcine NPY indicates that its mode of action is similar. These structural data can serve as a starting point in the design of new antiparasitic drugs.
- Department of Chemistry, University of Alberta, Edmonton, Alberta, T6G 2G2, Canada.
Organizational Affiliation: