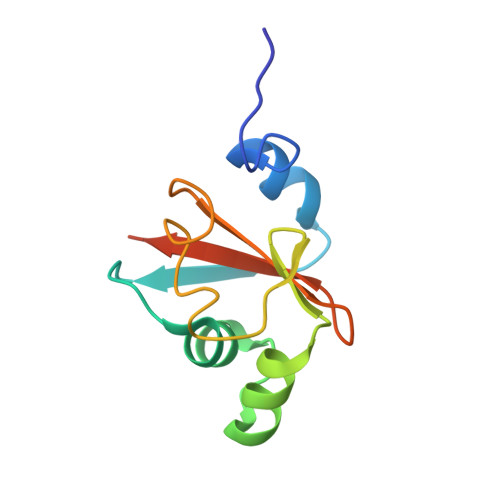
Structure and Dynamics of the Anticodon Arm Binding Domain of Bacillus stearothermophilus Tyrosyl-tRNA Synthetase
Guijarro, J.I., Pintar, A., Prochnicka-Chalufour, A., Guez, V., Gilquin, B., Bedouelle, H., Delepierre, M.(2002) Structure 10: 311-317
- PubMed: 12005430
- DOI: https://doi.org/10.1016/s0969-2126(02)00699-8
- Primary Citation of Related Structures:
1JH3 - PubMed Abstract:
The structure of a recombinant protein, TyrRS(delta4), corresponding to the anticodon arm binding domain of Bacillus stearothermophilus tyrosyl-tRNA synthetase, has been solved, and its dynamics have been studied by nuclear magnetic resonance (NMR). It is the first structure described for such a domain of a tyrosyl-tRNA synthetase. It consists of a five-stranded beta sheet, packed against two alpha helices on one side and one alpha helix on the other side. A large part of the domain is structurally similar to other functionally unrelated RNA binding proteins. The basic residues known to be essential for tRNA binding and charging are exposed to the solvent on the same face of the molecule. The structure of TyrRS(delta4), together with previous mutagenesis data, allows one to delineate the region of interaction with tRNATyr.
- Unité de RMN des Biomolécules, CNRS URA 2185, Institut Pasteur, Paris, France.
Organizational Affiliation: