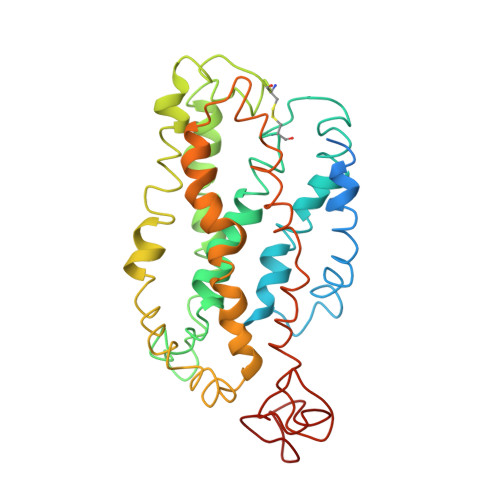
Studies on the structure of the G-protein-coupled receptor rhodopsin including the putative G-protein binding site in unactivated and activated forms.
Yeagle, P.L., Choi, G., Albert, A.D.(2001) Biochemistry 40: 11932-11937
- PubMed: 11570894
- DOI: https://doi.org/10.1021/bi015543f
- Primary Citation of Related Structures:
1JFP - PubMed Abstract:
Activation of G-protein coupled receptors (GPCR) is not yet understood. A recent structure showed most of rhodopsin in the ground (not activated) state of the GPCR, but the cytoplasmic face, which couples to the G protein in signal transduction, was not well-defined. We have determined an experimental three-dimensional structure for rhodopsin in the unactivated state, which shows good agreement with the crystal structure in the transmembrane domain. This new structure defines the cytoplasmic face of rhodopsin. The G-protein binding site can be mapped. The same experimental approach yields a preliminary structure of the cytoplasmic face in the activated (metarhodopsin II) receptor. Differences between the two structures suggest how the receptor is activated to couple with transducin.
- Department of Molecular and Cell Biology, University of Connecticut, Storrs, Connecticut 06269, USA. yeagle@uconnvm.uconn.edu
Organizational Affiliation: