Average Crystal Structure of (Pro-Pro-Gly)9 at 1.0 angstroms Resolution
Hongo, C., Nagarajan, V., Noguchi, K., Kamitori, S., Okuyama, K., Tanaka, Y., Nishino, N.(2001) Polym J 33: 812-818
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2001) Polym J 33: 812-818
Find similar proteins by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| COLLAGEN TRIPLE HELIX | 7 | N/A | Mutation(s): 0 | 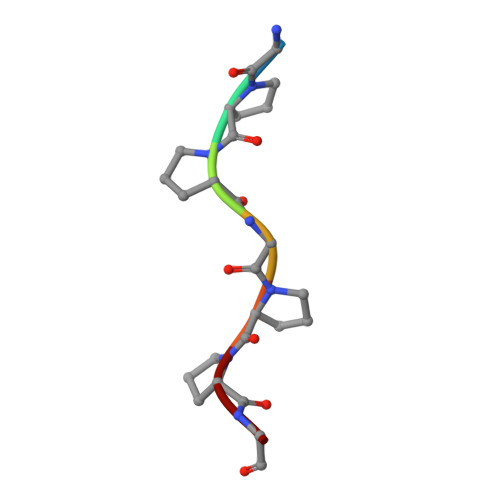 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| COLLAGEN TRIPLE HELIX | 7 | N/A | Mutation(s): 0 | 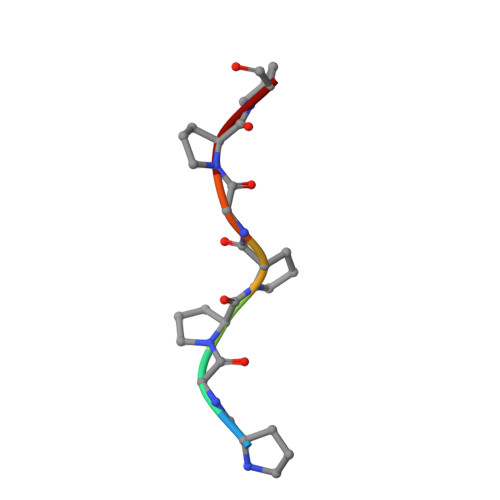 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 26.821 | α = 90 |
| b = 26.333 | β = 90 |
| c = 20.246 | γ = 90 |
| Software Name | Purpose |
|---|---|
| TEXSAN | data collection |
| teXsan | data reduction |
| X-PLOR | model building |
| SHELXL-97 | refinement |
| TEXSAN | data scaling |
| X-PLOR | phasing |