A new cytokine-receptor binding mode revealed by the crystal structure of the IL-1 receptor with an antagonist.
Schreuder, H., Tardif, C., Trump-Kallmeyer, S., Soffientini, A., Sarubbi, E., Akeson, A., Bowlin, T., Yanofsky, S., Barrett, R.W.(1997) Nature 386: 194-200
- PubMed: 9062194
- DOI: https://doi.org/10.1038/386194a0
- Primary Citation of Related Structures:
1IRA - PubMed Abstract:
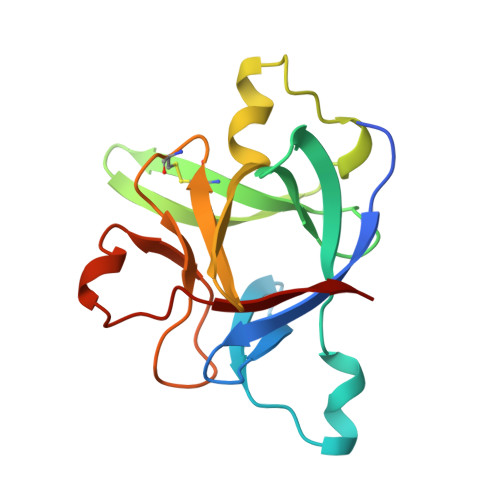
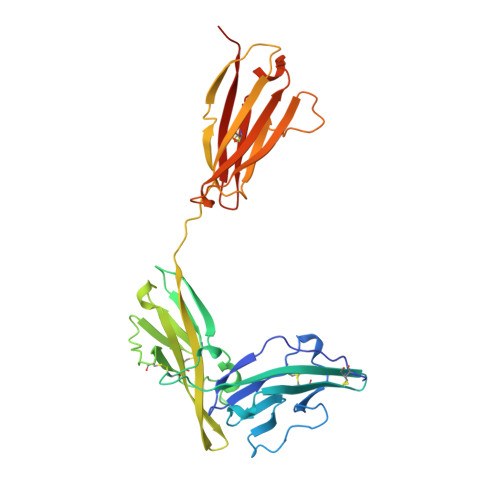
Inflammation, regardless of whether it is provoked by infection or by tissue damage, starts with the activation of macrophages which initiate a cascade of inflammatory responses by producing the cytokines interleukin-1 (IL-1) and tumour necrosis factor-alpha (ref. 1). Three naturally occurring ligands for the IL-1 receptor (IL1R) exist: the agonists IL-1alpha and IL-1beta and the IL-1-receptor antagonist IL1RA (ref. 2). IL-1 is the only cytokine for which a naturally occurring antagonist is known. Here we describe the crystal structure at 2.7 A resolution of the soluble extracellular part of type-I IL1R complexed with IL1RA. The receptor consists of three immunoglobulin-like domains. Domains 1 and 2 are tightly linked, but domain three is completely separate and connected by a flexible linker. Residues of all three domains contact the antagonist and include the five critical IL1RA residues which were identified by site-directed mutagenesis. A region that is important for biological function in IL-1beta, the 'receptor trigger site' is not in direct contact with the receptor in the IL1RA complex. Modelling studies suggest that this IL-1beta trigger site might induce a movement of domain 3.
- Marion Merrell Dow Research Institute, Strasbourg, France.
Organizational Affiliation: