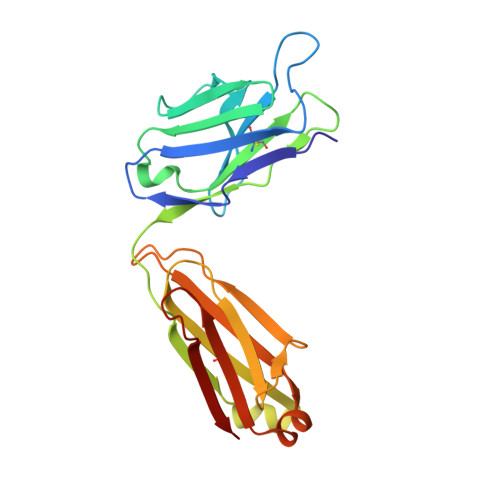
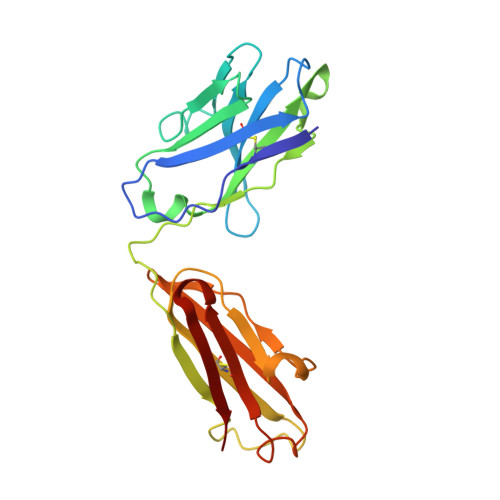
26-10 Fab-digoxin complex: affinity and specificity due to surface complementarity.
Jeffrey, P.D., Strong, R.K., Sieker, L.C., Chang, C.Y., Campbell, R.L., Petsko, G.A., Haber, E., Margolies, M.N., Sheriff, S.(1993) Proc Natl Acad Sci U S A 90: 10310-10314
- PubMed: 8234291
- DOI: https://doi.org/10.1073/pnas.90.21.10310
- Primary Citation of Related Structures:
1IGI, 1IGJ - PubMed Abstract:
We have determined the three-dimensional structures of the antigen-binding fragment of the anti-digoxin monoclonal antibody 26-10 in the uncomplexed state at 2.7 A resolution and as a complex with digoxin at 2.5 A resolution. Neither the antibody nor digoxin undergoes any significant conformational changes upon forming the complex. Digoxin interacts primarily with the antibody heavy chain and is oriented such that the carbohydrate groups are exposed to solvent and the lactone ring is buried in a deep pocket at the bottom of the combining site. Despite extensive interactions between antibody and antigen, no hydrogen bonds or salt links are formed between 26-10 and digoxin. Thus the 26-10-digoxin complex is unique among the known three-dimensional structures of antibody-antigen complexes in that specificity and high affinity arise primarily from shape complementarity.
- Bristol-Myers Squibb Pharmaceutical Research Institute, Princeton, NJ 08543-4000.
Organizational Affiliation: