Crystal structure of proteolytic fragments of the redox-sensitive Hsp33 with constitutive chaperone activity
Kim, S.J., Jeong, D.G., Chi, S.W., Lee, J.S., Ryu, S.E.(2001) Nat Struct Biol 8: 459-466
- PubMed: 11323724
- DOI: https://doi.org/10.1038/87639
- Primary Citation of Related Structures:
1I7F - PubMed Abstract:
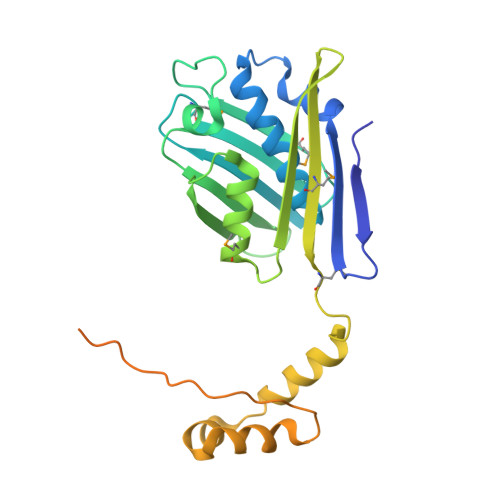
Heat shock protein 33 (Hsp33) inhibits aggregation of partially denatured proteins during oxidative stress. The chaperone activity of Hsp33 is unique among heat shock proteins because the activity is reversibly regulated by cellular redox status. We report here the crystal structure of the N-terminal region of Hsp33 fragments with constitutive chaperone activity. The structure reveals that the N-terminal portion of Hsp33 forms a tightly associated dimer formed by a domain crossover. A concave groove on the dimeric surface contains an elongated hydrophobic patch that could potentially bind denatured protein substrates. The termini of the subunits are located near the hydrophobic patch, indicating that the cleaved C-terminal domain may shield the hydrophobic patch in an inactive state. Two of the four conserved zinc-coordinating cysteines are in the end of the N-terminal domain, and the other two are in the cleaved C-terminal domain. The structural information and subsequent biochemical characterizations suggest that the redox switch of Hsp33 occurs by a reversible dissociation of the C-terminal regulatory domain through oxidation of zinc-coordinating cysteines and zinc release.
- Center for Cellular Switch Protein Structure, Korea Research Institute of Bioscience and Biotechnology, P. O. Box 115, Yusong, Taejon 305-600, South Korea.
Organizational Affiliation: