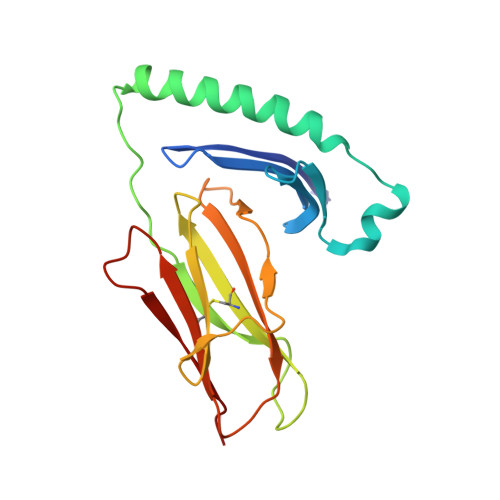
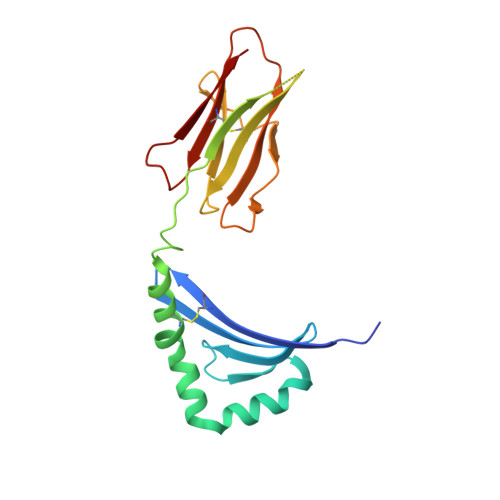
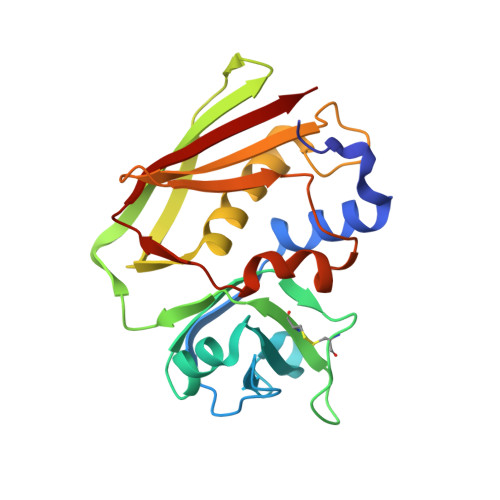
Crystal Structure of a Superantigen Bound to MHC Class II Displays Zinc and Peptide Dependence
Petersson, K., Hakansson, M., Nilsson, H., Forsberg, G., Svensson, L.A., Liljas, A., Walse, B.(2001) EMBO J 20: 3306-3312
- PubMed: 11432818
- DOI: https://doi.org/10.1093/emboj/20.13.3306
- Primary Citation of Related Structures:
1HXY - PubMed Abstract:
The three-dimensional structure of a bacterial superantigen, Staphylococcus aureus enterotoxin H (SEH), bound to human major histocompatibility complex (MHC) class II (HLA-DR1) has been determined by X-ray crystallography to 2.6 A resolution (1HXY). The superantigen binds on top of HLA-DR1 in a completely different way from earlier co-crystallized superantigens from S.aureus. SEH interacts with high affinity through a zinc ion with the beta1 chain of HLA-DR1 and also with the peptide presented by HLA-DR1. The structure suggests that all superantigens interacting with MHC class II in a zinc-dependent manner present the superantigen in a common way. This suggests a new model for ternary complex formation with the T-cell receptor (TCR), in which a contact between the TCR and the MHC class II is unlikely.
- Molecular Biophysics, Centre for Chemistry and Chemical Engineering, Lund University, PO Box 124, S-221 00 Lund, Sweden.
Organizational Affiliation: