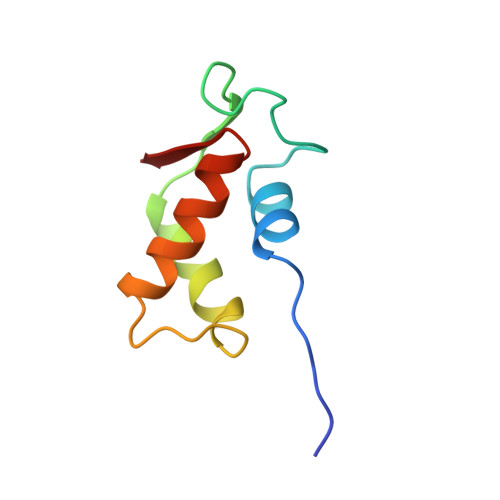
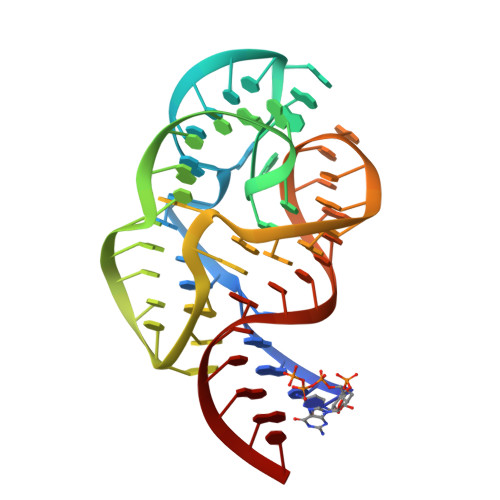
A Compact RNA Tertiary Structure Contains a Buried Backbone-K+ Complex
Conn, G.L., Gittis, A.G., Lattman, E.E., Misra, V.K., Draper, D.E.(2002) J Mol Biology 318: 963
- PubMed: 12054794
- DOI: https://doi.org/10.1016/S0022-2836(02)00147-X
- Primary Citation of Related Structures:
1HC8 - PubMed Abstract:
The structure of a 58 nucleotide ribosomal RNA fragment buries several phosphate groups of a hairpin loop within a large tertiary core. During refinement of an X-ray crystal structure containing this RNA, a potassium ion was found to be contacted by six oxygen atoms from the buried phosphate groups; the ion is contained completely within the solvent-accessible surface of the RNA. The electrostatic potential at the ion chelation site is unusually large, and more than compensates for the substantial energetic penalties associated with partial dehydration of the ion and displacement of delocalized ions. The very large predicted binding free energy, approximately -30 kcal/mol, implies that the site must be occupied for the RNA to fold. These findings agree with previous studies of the ion-dependent folding of tertiary structure in this RNA, which concluded that a monovalent ion was bound in a partially dehydrated environment where Mg2+ could not easily compete for binding. By compensating the unfavorable free energy of buried phosphate groups with a chelated ion, the RNA is able to create a larger and more complex tertiary fold than would be possible otherwise.
- Department of Chemistry, The Johns Hopkins University, 3400 North Charles Street, Baltimore, MD 21218, USA.
Organizational Affiliation: