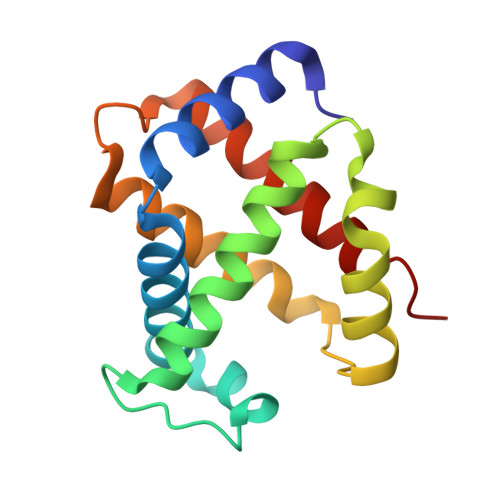
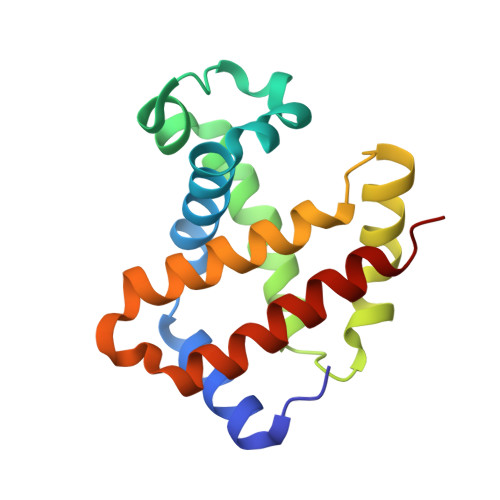
Crystal Structure of T State Haemoglobin with Oxygen Bound at All Four Haems.
Paoli, M., Liddington, R., Tame, J., Wilkinson, A., Dodson, G.(1996) J Mol Biology 256: 775
- PubMed: 8642597
- DOI: https://doi.org/10.1006/jmbi.1996.0124
- Primary Citation of Related Structures:
1GZX - PubMed Abstract:
The cooperative binding of oxygen by haemoglobin results from restraints on ligand binding in the T state. The unfavourable interactions made by the ligands at the haems destabilise the T state and favour the high affinity R state. The T <==> R equilibrium leads, in the presence of a ligand, to a rapid increase in the R state population and therefore generates cooperative binding. There is now considerable understanding of this phenomenon, but the interactions that reduce ligand affinity in the T state have not yet been fully explored, owing to the difficulties in preparing T state haemoglobin crystals in which all the subunits are oxygenated. A protocol has been developed to oxygenate deoxy T state adult human haemoglobin (HbA) crystals in air at 4 C at all four haems without significant loss of crystalline order. The X-ray crystal structure, determined to 2.1 A spacing, shows significant changes in the alpha and beta haem pockets as well as changes at the alpha(1)beta(2) interface in the direction of the R quaternary structure. Most of the shifts and deviations from deoxy T state HbA are similar to, but larger than, those previously observed in the T state met and other partially liganded T state forms. They provide clear evidence of haem-haem interaction in the T state.
- Department of Chemistry, University of York, Heslington, York, UK.
Organizational Affiliation: