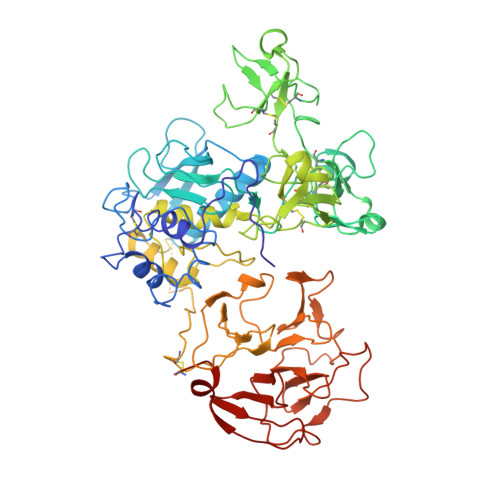
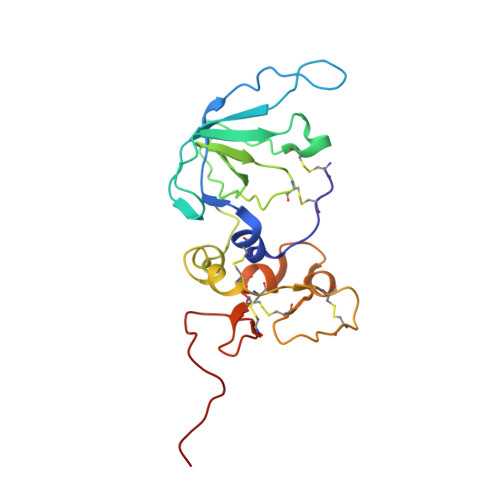
Structural Insight Into the Complex Formation of Latent Matrix Metalloproteinase 2 with Tissue Inhibitor of Metalloproteinase 2
Morgunova, E., Tuuttila, A., Bergmann, U., Tryggvason, K.(2002) Proc Natl Acad Sci U S A 99: 7414
- PubMed: 12032297
- DOI: https://doi.org/10.1073/pnas.102185399
- Primary Citation of Related Structures:
1GXD - PubMed Abstract:
Matrix metalloproteinases (MMPs) are a family of multidomain enzymes involved in the physiological degradation of connective tissue, as well as in pathological states such as tumor invasion and arthritis. Apart from transcriptional regulation, MMPs are controlled by proenzyme activation and a class of specific tissue inhibitors of metalloproteinases (TIMPs) that bind to the catalytic site. TIMP-2 is a potent inhibitor of MMPs, but it has also been implicated in a unique cell surface activation mechanism of latent MMP-2/gelatinase A/type IV collagenase (proMMP-2), through its binding to the hemopexin domain of proMMP-2 on the one hand and to a membrane-type MMP activator on the other. The present crystal structure of the human proMMP-2/TIMP-2 complex reveals an interaction between the hemopexin domain of proMMP-2 and the C-terminal domain of TIMP-2, leaving the catalytic site of MMP-2 and the inhibitory site of TIMP-2 distant and spatially isolated. The interfacial contact of these two proteins is characterized by two distinct binding regions composed of alternating hydrophobic and hydrophilic interactions. This unique structure provides information for how specificity for noninhibitory MMP/TIMP complex formation is achieved.
- Division of Matrix Biology, Department of Medical Biochemistry and Biophysics, Karolinska Institutet, S-171 77 Stockholm, Sweden.
Organizational Affiliation: