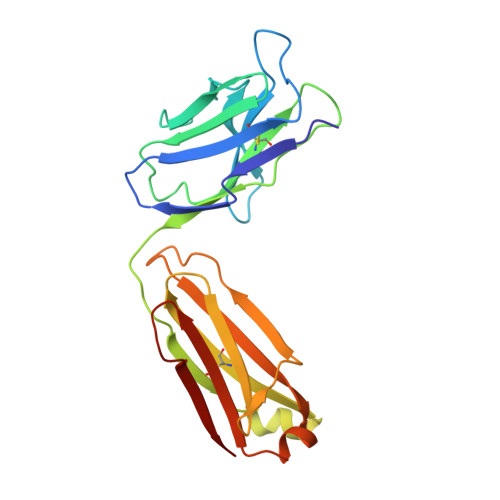
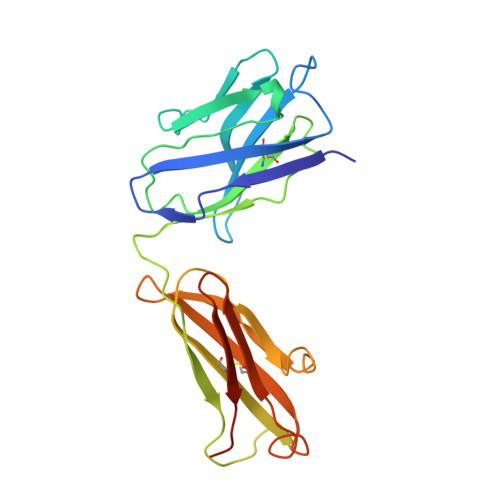
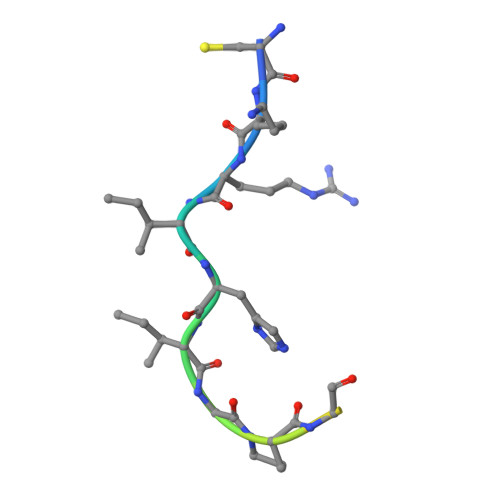
Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen.
Rini, J.M., Stanfield, R.L., Stura, E.A., Salinas, P.A., Profy, A.T., Wilson, I.A.(1993) Proc Natl Acad Sci U S A 90: 6325-6329
- PubMed: 8327513
- DOI: https://doi.org/10.1073/pnas.90.13.6325
- Primary Citation of Related Structures:
1GGI - PubMed Abstract:
The crystal structure of the Fab fragment of a human immunodeficiency virus type 1 (HIV-1) neutralizing monoclonal antibody Fab has been determined at 2.8 A resolution in complex with a linear 16-residue peptide from the third hypervariable region (V3) of gp120. The first 9 residues of the peptide are ordered in the electron density maps, and their conformation is in partial agreement with the beta-strand-type II beta-turn structure predicted for this portion of the V3 loop. Notably, several of the peptide residues that are well conserved among different HIV-1 isolates contact a nonpolar 25-A-long groove in the antibody-combining site. The largely extended structure of the peptide differs from the beta-turns seen as the primary determinants in other published anti-peptide Fab structures. Analysis of the specific Fab-peptide interactions only partially explains the MN isolate specificity shown by this antibody.
- Department of Molecular Biology, Scripps Research Institute, La Jolla, CA 92037.
Organizational Affiliation: