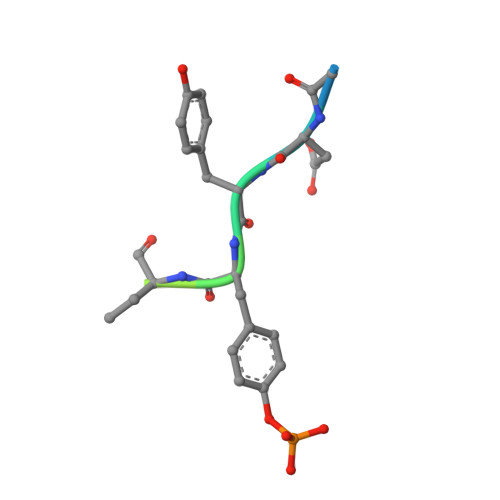
Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B.
Salmeen, A., Andersen, J.N., Myers, M.P., Tonks, N.K., Barford, D.(2000) Mol Cell 6: 1401-1412
- PubMed: 11163213
- DOI: https://doi.org/10.1016/s1097-2765(00)00137-4
- Primary Citation of Related Structures:
1G1F, 1G1G, 1G1H - PubMed Abstract:
The protein tyrosine phosphatase PTP1B is responsible for negatively regulating insulin signaling by dephosphorylating the phosphotyrosine residues of the insulin receptor kinase (IRK) activation segment. Here, by integrating crystallographic, kinetic, and PTP1B peptide binding studies, we define the molecular specificity of this reaction. Extensive interactions are formed between PTP1B and the IRK sequence encompassing the tandem pTyr residues at 1162 and 1163 such that pTyr-1162 is selected at the catalytic site and pTyr-1163 is located within an adjacent pTyr recognition site. This selectivity is attributed to the 70-fold greater affinity for tandem pTyr-containing peptides relative to mono-pTyr peptides and predicts a hierarchical dephosphorylation process. Many elements of the PTP1B-IRK interaction are unique to PTP1B, indicating that it may be feasible to generate specific, small molecule inhibitors of this interaction to treat diabetes and obesity.
- Laboratory of Molecular Biophysics, University of Oxford, South Parks Road, Oxford, OX1 3QU, United Kingdom.
Organizational Affiliation: