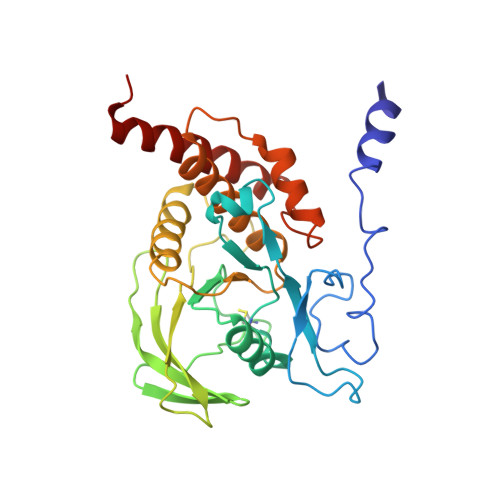
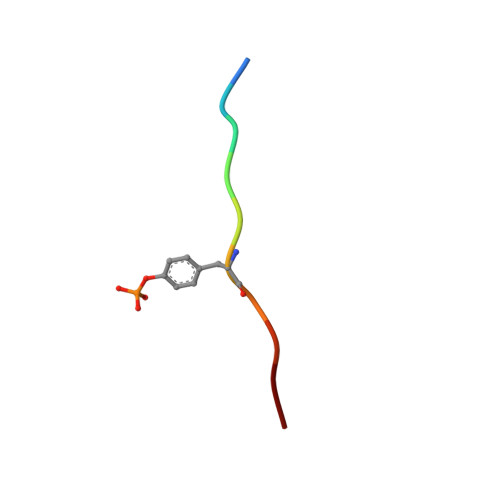
Structural basis for substrate specificity of protein-tyrosine phosphatase SHP-1.
Yang, J., Cheng, Z., Niu, T., Liang, X., Zhao, Z.J., Zhou, G.W.(2000) J Biological Chem 275: 4066-4071
- PubMed: 10660565
- DOI: https://doi.org/10.1074/jbc.275.6.4066
- Primary Citation of Related Structures:
1FPR - PubMed Abstract:
The substrate specificity of the catalytic domain of SHP-1, an important regulator in the proliferation and development of hematopoietic cells, is critical for understanding the physiological functions of SHP-1. Here we report the crystal structures of the catalytic domain of SHP-1 complexed with two peptide substrates derived from SIRPalpha, a member of the signal-regulatory proteins. We show that the variable beta5-loop-beta6 motif confers SHP-1 substrate specificity at the P-4 and further N-terminal subpockets. We also observe a novel residue shift at P-2, the highly conserved subpocket in protein- tyrosine phosphatases. Our observations provide new insight into the substrate specificity of SHP-1.
- Program in Molecular Medicine, University of Massachusetts Medical School, Worcester, Massachusetts 01605, USA.
Organizational Affiliation: