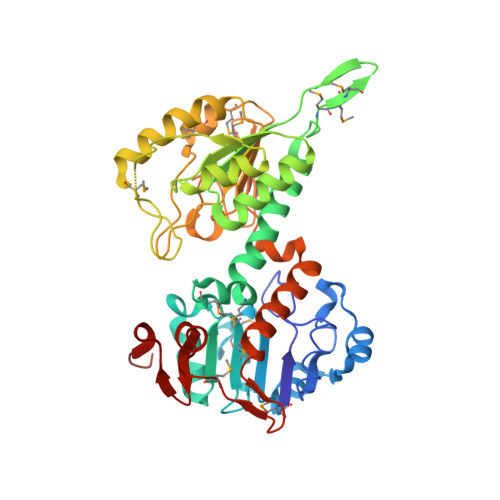
Protein-protein recognition, hydride transfer and proton pumping in the transhydrogenase complex.
Buckley, P.A., Baz Jackson, J., Schneider, T., White, S.A., Rice, D.W., Baker, P.J.(2000) Structure 8: 809-815
- PubMed: 10997900
- DOI: https://doi.org/10.1016/s0969-2126(00)00171-4
- Primary Citation of Related Structures:
1F8G - PubMed Abstract:
Membrane-bound ion pumps are involved in metabolic regulation, osmoregulation, cell signalling, nerve transmission and energy transduction. How the ion electrochemical gradient interacts with the scalar chemistry and how the catalytic machinery is gated to ensure high coupling efficiency are fundamental to the mechanism of action of such pumps. Transhydrogenase is a conformationally coupled proton pump linking a proton gradient to the redox reaction between NAD(H) and NADP(H). The enzyme has three components; dI binds NAD(H), dII spans the membrane and dIII binds NADP(H). The first crystal structure of a transhydrogenase dI component (from Rhodospirillum rubrum) has been determined at 2.0 A resolution. The monomer comprises two domains. Both are involved in dimer formation, and one has a Rossmann fold that binds NAD+ in a novel mode. The two domains can adopt different conformations. In the most closed conformation, the nicotinamide ring is expelled from the cleft between the two domains and is exposed on the outside of the protein. In this conformation it is possible to dock the structure of dI/NAD+ with that of a dIII/NADP+ complex to provide the first insights into the molecular basis of the hydride-transfer step. Analysis of the model of the dI/dIII complex identifies residues potentially involved in dI/dIII interaction and shows how domain motion in dI results in a shift in position of the nicotinamide ring of NAD+. We propose that this movement is responsible for switching between the forbidden and allowed states for hydride transfer during proton pumping.
- Department of Molecular Biology and Biotechnology, University of Sheffield, Western Bank, UK.
Organizational Affiliation: