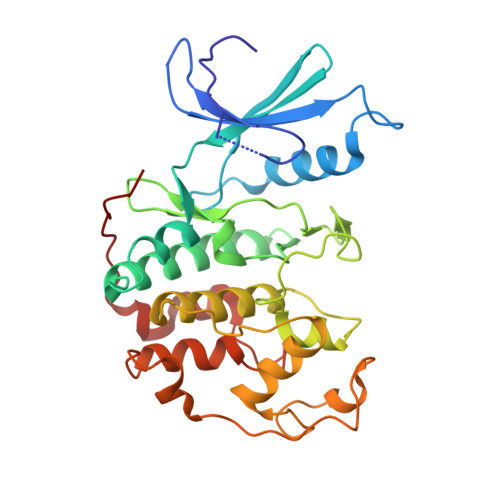
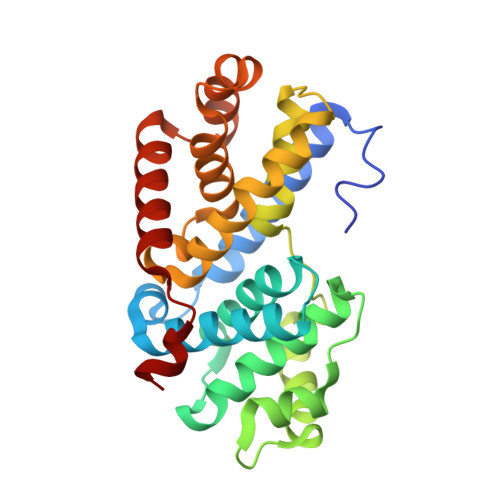
Crystal structure of a gamma-herpesvirus cyclin-cdk complex.
Card, G.L., Knowles, P., Laman, H., Jones, N., McDonald, N.Q.(2000) EMBO J 19: 2877-2888
- PubMed: 10856233
- DOI: https://doi.org/10.1093/emboj/19.12.2877
- Primary Citation of Related Structures:
1F5Q - PubMed Abstract:
Several gamma-herpesviruses encode proteins related to the mammalian cyclins, regulatory subunits of cyclin-dependent kinases (cdks) essential for cell cycle progression. We report a 2.5 A crystal structure of a full-length oncogenic viral cyclin from gamma-herpesvirus 68 complexed with cdk2. The viral cyclin binds cdk2 with an orientation different from cyclin A and makes several novel interactions at the interface, yet it activates cdk2 by triggering conformational changes similar to cyclin A. Sequences within the viral cyclin N-terminus lock part of the cdk2 T-loop within the core of the complex. These sequences and others are conserved amongst the viral and cellular D-type cyclins, suggesting that this structure has wider implications for other cyclin-cdk complexes. The observed resistance of this viral cyclin-cdk complex to inhibition by the p27(KIP:) cdk inhibitor is explained by sequence and conformational variation in the cyclin rendering the p27(KIP:)-binding site on the cyclin subunit non-functional.
- Structural Biology and Gene Expression Laboratories, Imperial Cancer Research Fund, 44 Lincoln's Inn Fields, Holborn, London WC2A 3PX, UK.
Organizational Affiliation: