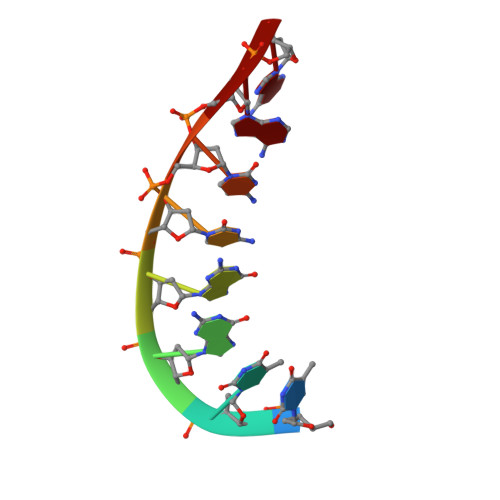
A high-resolution structure of a DNA-chromomycin-Co(II) complex determined from pseudocontact shifts in nuclear magnetic resonance.
Gochin, M.(2000) Structure 8: 441-452
- PubMed: 10801486
- DOI: https://doi.org/10.1016/s0969-2126(00)00124-6
- Primary Citation of Related Structures:
1EKH, 1EKI - PubMed Abstract:
The drug chromomycin-A(3) binds to the minor groove of DNA and requires a divalent metal ion for complex formation. (1)H, (31)P and (13)C pseudocontact shifts occurring in the presence of a tightly bound divalent cobalt ion in the complex between d(TTGGCCAA)(2) and chromomycin-A(3) have been used to determine the structure of the complex. The accuracy of the structure was verified by validation with nuclear Overhauser enhancements (NOEs) and J-coupling constants not used in the structure calculation. The final structure was determined to 0.7 A resolution. The structure was compared with a structure obtained in an earlier study using NOEs, in order to assess the accuracy of NOEs in giving global structural information for a DNA complex. Although some basic features of the structures agreed, they differed substantially in the fine structural details and in the DNA axis curvature generated by the drug. The distortion of base-pair planarity that was observed in the NOE structure was not seen in our structure. Differences in drug orientation and hydrogen bonding also occurred. The curvature and elongation of the DNA that was obtained previously was not found to occur in our study. The use of pseudocontact shifts has enabled us to obtain a high-precision global structure of the chromomycin-DNA complex, which provides an accurate template on which to consider targeting minor groove binding drugs. The effect of such binding is not propagated far along the helix but is restricted to a local kink in the axis that reverts to its original direction within four base pairs.
- Department of Microbiology, University of the Pacific School of Dentistry, San Francisco, CA 94115, USA. miriam@picasso. ucsf.edu
Organizational Affiliation: