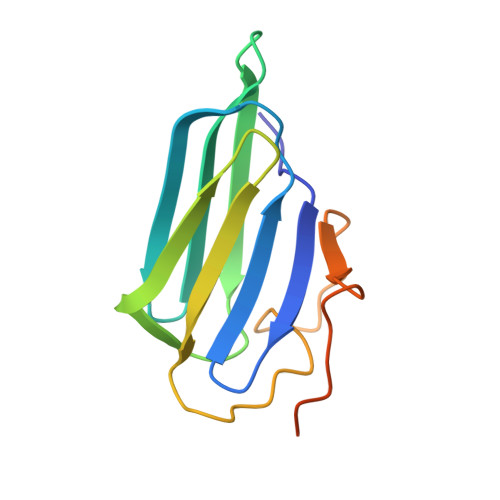
Crystal structure and activity of human p23, a heat shock protein 90 co-chaperone.
Weaver, A.J., Sullivan, W.P., Felts, S.J., Owen, B.A., Toft, D.O.(2000) J Biological Chem 275: 23045-23052
- PubMed: 10811660
- DOI: https://doi.org/10.1074/jbc.M003410200
- Primary Citation of Related Structures:
1EJF - PubMed Abstract:
p23 is a co-chaperone for the heat shock protein, hsp90. This protein binds hsp90 and participates in the folding of a number of cell regulatory proteins, but its activities are still unclear. We have solved a crystal structure of human p23 lacking 35 residues at the COOH terminus. The structure reveals a disulfide-linked dimer with each subunit containing eight beta-strands in a compact antiparallel beta-sandwich fold. In solution, however, p23 is primarily monomeric and the dimer appears to be a minor component. Conserved residues are clustered on one face of the monomer and define a putative surface region and binding pocket for interaction(s) with hsp90 or protein substrates. p23 contains a COOH-terminal tail that is apparently less structured and is unresolved in the crystal structure. This tail is not needed for the binding of p23 to hsp90 or to complexes with the progesterone receptor. However, the tail is necessary for optimum active chaperoning of the progesterone receptor, as well as the passive chaperoning activity of p23 in assays measuring inhibition of heat-induced protein aggregation.
- hkl Research, Inc., Ithaca, New York 14853, USA.
Organizational Affiliation: