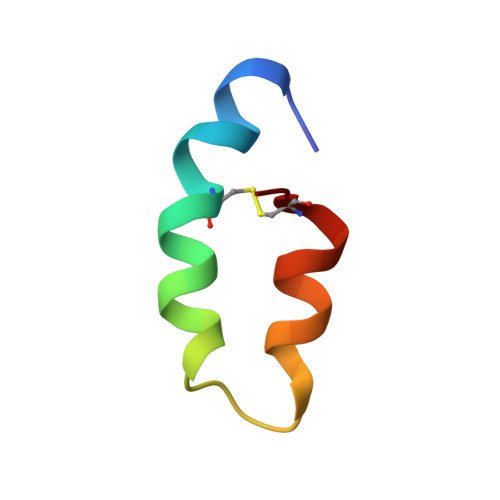
Three-dimensional structure of ectatomin from Ectatomma tuberculatum ant venom.
Nolde, D.E., Sobol, A.G., Pluzhnikov, K.A., Grishin, E.V., Arseniev, A.S.(1995) J Biomol NMR 5: 1-13
- PubMed: 7881269
- DOI: https://doi.org/10.1007/BF00227465
- Primary Citation of Related Structures:
1ECI - PubMed Abstract:
Two-dimensional 1H NMR techniques were used to determine the spatial structure of ectatomin, a toxin from the venom of the ant Ectatomma tuberculatum. Nearly complete proton resonance assignments for two chains of ectatomin (37 and 34 amino acid residues, respectively) were obtained using 2D TOCSY, DQF-COSY and NOESY experiments. The cross-peak volumes in NOESY spectra were used to define the local structure of the protein and generate accurate proton-proton distance constraints employing the MARDIGRAS program. Disulfide bonds were located by analyzing the global fold of ectatomin, calculated with the distance geometry program DIANA. These data, combined with data on the rate of exchange of amide protons with deuterium, were used to obtain a final set of 20 structures by DIANA. These structures were refined by unrestrained energy minimization using the CHARMm program. The resulting rms deviations over 20 structures (excluding the mobile N- and c-termini of each chain) are 0.75 A for backbone heavy atoms, and 1.25 A for all heavy atoms. The conformations of the two chains are similar. Each chain consists of two alpha-helices and a hinge region of four residues; this forms a hairpin structure which is stabilized by disulfide bridges. The hinge regions of the two chains are connected together by a third disulfide bridge. Thus, ectatomin forms a four-alpha-helical bundle structure.
- Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow.
Organizational Affiliation: