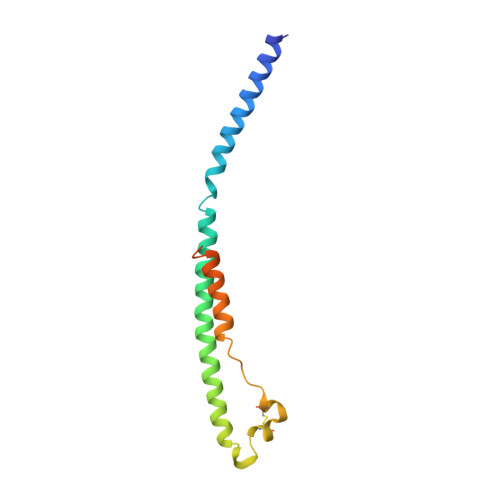
Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain.
Weissenhorn, W., Carfi, A., Lee, K.H., Skehel, J.J., Wiley, D.C.(1998) Mol Cell 2: 605-616
- PubMed: 9844633
- DOI: https://doi.org/10.1016/s1097-2765(00)80159-8
- Primary Citation of Related Structures:
1EBO - PubMed Abstract:
We have determined the structure of GP2 from the Ebola virus membrane fusion glycoprotein by X-ray crystallography. The molecule contains a central triple-stranded coiled coil followed by a disulfide-bonded loop homologous to an immunosuppressive sequence in retroviral glycoproteins, which reverses the chain direction and connects to an alpha helix packed antiparallel to the core helices. The structure suggests that fusion peptides near the N termini form disulfide-bonded loops at one end of the molecule and that the C-terminal membrane anchors are at the same end. In this conformation, GP2 could both bridge two membranes and facilitate their apposition to initiate membrane fusion. We also find a heptad irregularity like that in low-pH-induced influenza HA2 and a solvent ion trapped in a coiled coil like that in retroviral TMs.
- Laboratory of Molecular Medicine, Howard Hughes Medical Institute, Children's Hospital, Boston, Massachusetts 02115, USA.
Organizational Affiliation: