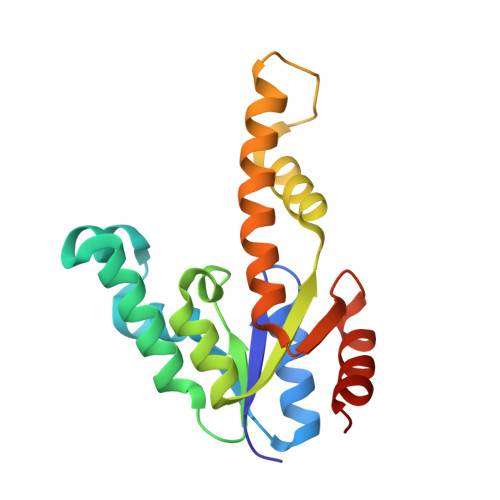
Biochemical and X-Ray Crystallographic Studies on Shikimate Kinase: The Important Structural Role of the P-Loop Lysine
Krell, T., Maclean, J., Boam, D.J., Cooper, A., Resmini, M., Brocklehurst, K., Kelly, S.M., Price, N.C., Lapthorn, A.J., Coggins, J.R.(2001) Protein Sci 10: 1137
- PubMed: 11369852
- DOI: https://doi.org/10.1110/ps.52501
- Primary Citation of Related Structures:
1E6C - PubMed Abstract:
Shikimate kinase, despite low sequence identity, has been shown to be structurally a member of the nucleoside monophosphate (NMP) kinase family, which includes adenylate kinase. In this paper we have explored the roles of residues in the P-loop of shikimate kinase, which forms the binding site for nucleotides and is one of the most conserved structural features in proteins. In common with many members of the P-loop family, shikimate kinase contains a cysteine residue 2 amino acids upstream of the essential lysine residue; the side chains of these residues are shown to form an ion pair. The C13S mutant of shikimate kinase was found to be enzymatically active, whereas the K15M mutant was inactive. However, the latter mutant had both increased thermostability and affinity for ATP when compared to the wild-type enzyme. The structure of the K15M mutant protein has been determined at 1.8 A, and shows that the organization of the P-loop and flanking regions is heavily disturbed. This indicates that, besides its role in catalysis, the P-loop lysine also has an important structural role. The structure of the K15M mutant also reveals that the formation of an additional arginine/aspartate ion pair is the most likely reason for its increased thermostability. From studies of ligand binding it appears that, like adenylate kinase, shikimate kinase binds substrates randomly and in a synergistic fashion, indicating that the two enzymes have similar catalytic mechanisms.
- Division of Biochemistry and Molecular Biology, Institute of Biomedical and Life Sciences, University of Glasgow, Glasgow G12 8QQ, Scotland, UK.
Organizational Affiliation: