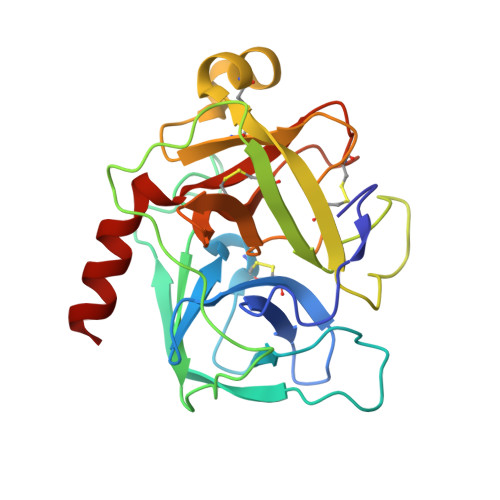
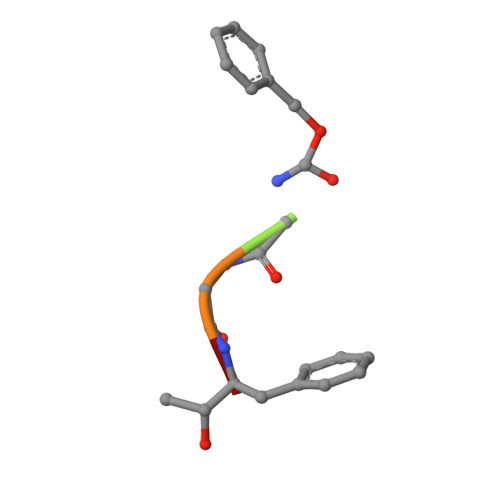
Crystal structure of delta-chymotrypsin bound to a peptidyl chloromethyl ketone inhibitor.
Mac Sweeney, A., Birrane, G., Walsh, M.A., O'Connell, T., Malthouse, J.P., Higgins, T.M.(2000) Acta Crystallogr D Biol Crystallogr 56: 280-286
- PubMed: 10713514
- DOI: https://doi.org/10.1107/s0907444999016583
- Primary Citation of Related Structures:
1DLK - PubMed Abstract:
Chymotrypsin is a member of the trypsin family of serine proteases and is one of the first proteins successfully studied by X-ray crystallography. It is secreted into the intestine as the inactive precursor chymotrypsinogen; four sequential cleavages of the peptide bonds following residues 13, 15, 146 and 148 occur to generate the active pi, delta, kappa and alpha forms of chymotrypsin. (13)C NMR has shown [O'Connell & Malthouse (1995). Biochem. J. 307, 353-359] that when the delta form of chymotrypsin is inhibited by 2-(13)C-enriched benzyloxycarbonylglycylglycylphenylalanyl chloromethane, a tetrahedral adduct is formed which is thought to be analogous to the tetrahedral intermediate formed during catalysis. This inhibitor complex has been crystallized as a dimer in space group P4(1)2(1)2. The structure has been refined at 2.14 A resolution to an R value of 21.2% (free R = 25.2%). Conformational differences between delta-chymotrypsin and chymotrypsinogen in the region of the flexible autolysis loop (residues 145-150) were observed. This is the first crystal structure of delta-chymotrypsin and includes two residues which are disordered in previous crystal structures of active chymotrypsin. A difference of 11.3 A(2) between the average B values of the monomers within the asymmetric unit is caused by lattice-disordering effects approximating to rotation of the molecules about a crystallographic screw axis. The substrate-binding mode of the inhibitor was similar to other chymotrypsin peptidyl inhibitor complexes, but this is the first published chymotrypsin structure in which the tetrahedral chloromethyl ketone transition-state analogue is observed. This structure is compared with that of a similar tetrahedral transition-state analogue which does not alkylate the active-site histidine residue.
- Department of Chemistry, NUI Galway, Galway, Ireland. macsweeney@biocfebs.unizh.ch
Organizational Affiliation: