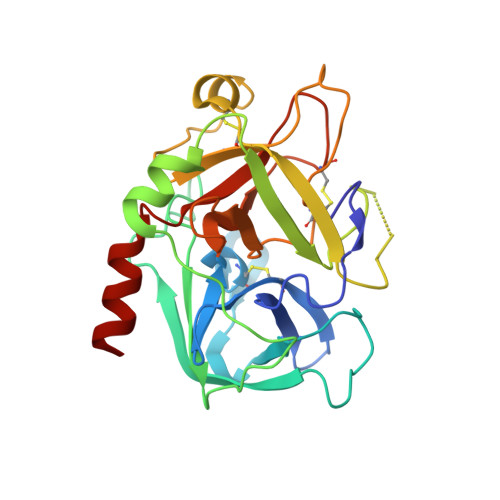
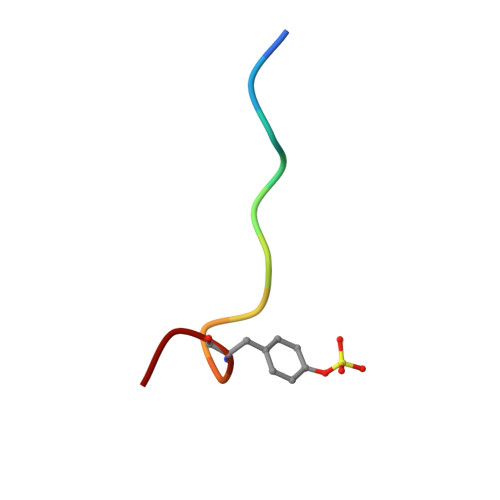
The crystal structures of human alpha-thrombin complexed with active site-directed diamino benzo[b]thiophene derivatives: a binding mode for a structurally novel class of inhibitors.
Chirgadze, N.Y., Sall, D.J., Briggs, S.L., Clawson, D.K., Zhang, M., Smith, G.F., Schevitz, R.W.(2000) Protein Sci 9: 29-36
- PubMed: 10739244
- DOI: https://doi.org/10.1110/ps.9.1.29
- Primary Citation of Related Structures:
1D3D, 1D3P, 1D3Q, 1D3T - PubMed Abstract:
The crystal structures of four active site-directed thrombin inhibitors, 1-4, in a complex with human alpha-thrombin have been determined and refined at up to 2.0 A resolution using X-ray crystallography. These compounds belong to a structurally novel family of inhibitors based on a 2,3-disubstituted benzo[b]thiophene structure. Compared to traditional active-site directed inhibitors, the X-ray crystal structures of these complexes reveal a novel binding mode. Unexpectedly, the lipophilic benzo[b]thiophene nucleus of the inhibitor appears to bind in the S1 specificity pocket. At the same time, the basic amine of the C-3 side chain of the inhibitor interacts with the mostly hydrophobic proximal, S2, and distal, S3, binding sites. The second, basic amine side chain at C-2 was found to point away from the active site, occupying a location between the S1 and S1' sites. Together, the aromatic rings of the C-2 and C-3 side chains sandwich the indole ring of Trp60D contained in the thrombin S2 insertion loop defined by the sequence "Tyr-Pro-Pro-Trp." [The thrombin residue numbering used in this study is equivalent to that reported for chymotrypsinogen (Hartley BS, Shotton DM, 1971, The enzymes, vol. 3. New York: Academic Press. pp 323-373).] In contrast to the binding mode of more classical thrombin inhibitors (D-Phe-Pro-Arg-H, NAPAP, Argatroban), this novel class of benzo[b]thiophene derivatives does not engage in hydrogen bond formation with Gly216 of the thrombin active site. A detailed analysis of the three-dimensional structures not only provides a clearer understanding of the interaction of these agents with thrombin, but forms a foundation for rational structure-based drug design. The use of the data from this study has led to the design of derivatives that are up to 2,900-fold more potent than the screening hit 1.
- Lilly Research Laboratories, Lilly Corporate Center, Indianapolis, Indiana 46285, USA. nyc@lilly.com
Organizational Affiliation: