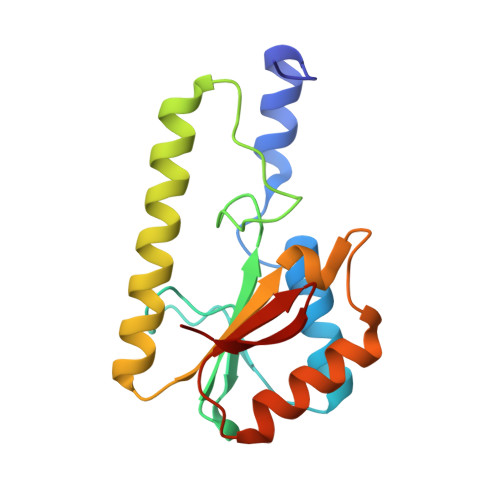
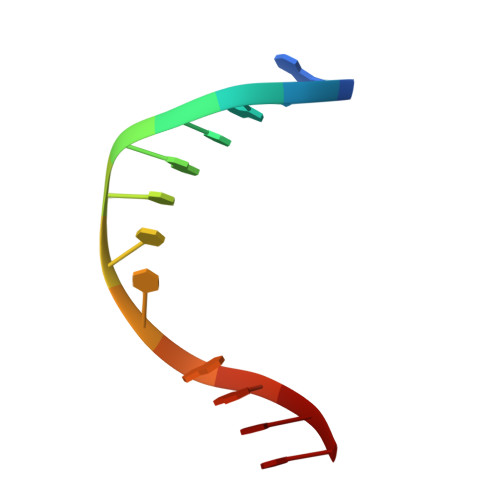
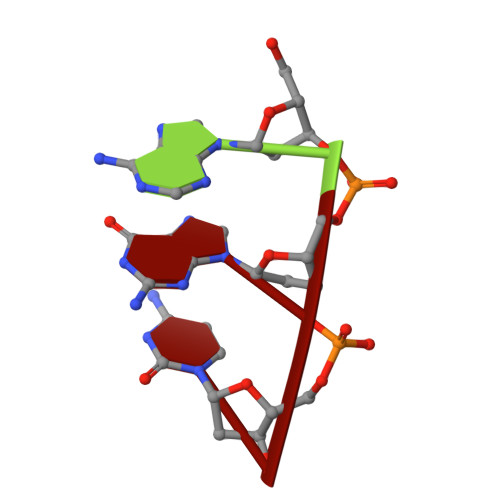
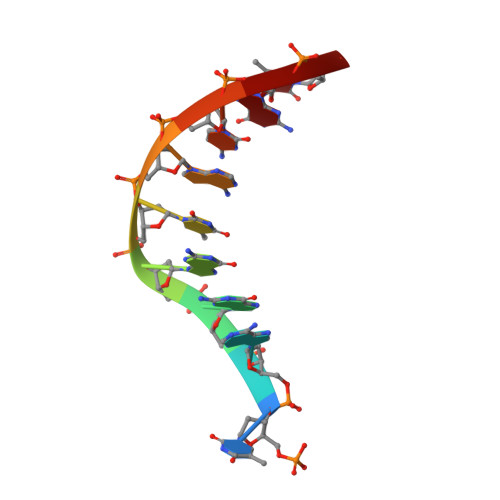
Recognition of a TG mismatch: the crystal structure of very short patch repair endonuclease in complex with a DNA duplex.
Tsutakawa, S.E., Jingami, H., Morikawa, K.(1999) Cell 99: 615-623
- PubMed: 10612397
- DOI: https://doi.org/10.1016/s0092-8674(00)81550-0
- Primary Citation of Related Structures:
1CW0 - PubMed Abstract:
The crystal structure of very short patch repair (Vsr) endonuclease, in complex with Mg2+ and with duplex DNA containing a TG mismatch, has been determined at 2.3 A resolution. In E. coli, the enzyme recognizes a TG mismatched base pair, generated after spontaneous deamination of methylated cytosines, and cleaves the phosphate backbone on the 5' side of the thymine. Extensive interactions between the DNA and the protein characterize a novel recognition mechanism, where three aromatic residues intercalate from the major groove into the DNA to strikingly deform the base pair stacking. With the presence of a cleaved DNA intermediate in the active center, the structure of the Vsr/DNA complex provides detailed insights into the catalytic mechanism for endonuclease activity.
- Department of Structural Biology, Biomolecular Engineering Research Institute, Suita, Osaka, Japan.
Organizational Affiliation: