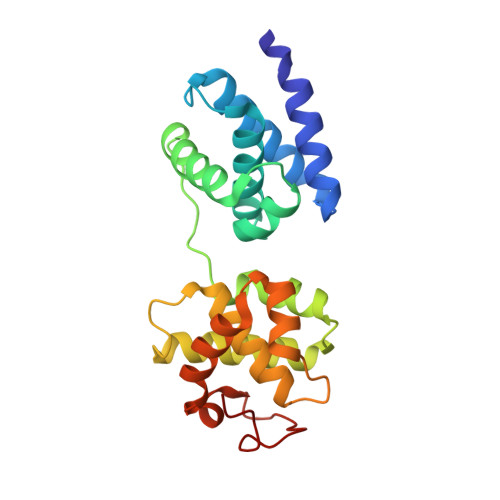
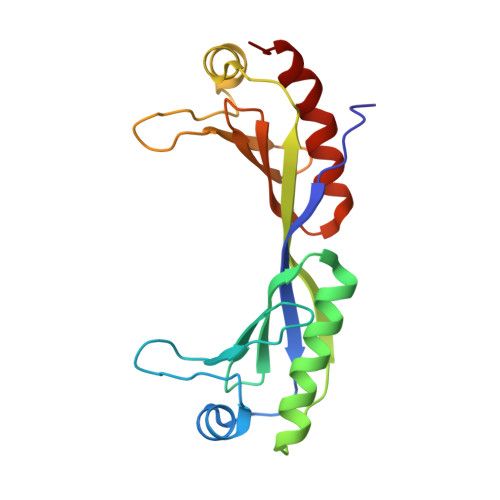
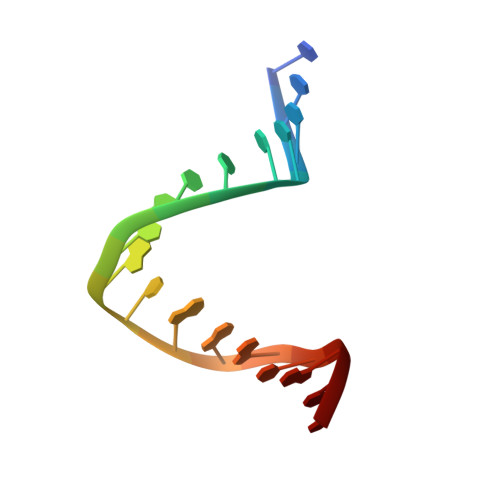
Structural basis of preinitiation complex assembly on human pol II promoters.
Tsai, F.T., Sigler, P.B.(2000) EMBO J 19: 25-36
- PubMed: 10619841
- DOI: https://doi.org/10.1093/emboj/19.1.25
- Primary Citation of Related Structures:
1C9B - PubMed Abstract:
Transcription initiation requires the assembly of a preinitiation complex (PIC), which is nucleated through binding of the TATA-box binding protein (TBP) to the promoter. Biochemical studies have shown, however, that TBP recognizes the TATA-box in both orientations and, therefore, cannot account for the directionality of PIC assembly. Transcription factor IIB (TFIIB) is essential for transcription initiation from RNA polymerase II promoters. Recent functional studies have identified a specific 7 bp TFIIB recognition element (BRE) immediately upstream of the TATA-box. We present here the 2.65 A resolution crystal structure of a human TFIIBc-TBPc complex bound to an idealized and extended adenovirus major late promoter. This structure now reveals that human TFIIBc binds to the promoter asymmetrically through base-specific contacts in the major groove upstream and in the minor groove downstream of the TATA-box. Binding of TFIIBc is, therefore, synergistic with TBPc requiring the distortion of the TATA-box. Thus, the newly described TFIIBc-DNA interface is likely to be a key determinant for the unidirectional assembly of a functional PIC.
- Department of Molecular Biophysics and Biochemistry and the Howard Hughes Medical Institute, Yale University, New Haven, CT 06511, USA.
Organizational Affiliation: