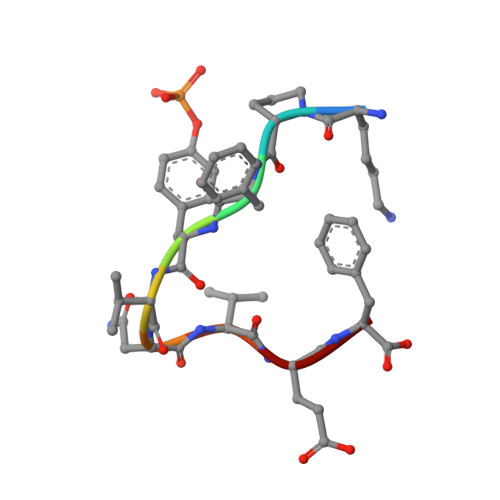
Structural and conformational requirements for high-affinity binding to the SH2 domain of Grb2(1).
Ettmayer, P., France, D., Gounarides, J., Jarosinski, M., Martin, M.S., Rondeau, J.M., Sabio, M., Topiol, S., Weidmann, B., Zurini, M., Bair, K.W.(1999) J Med Chem 42: 971-980
- PubMed: 10090780
- DOI: https://doi.org/10.1021/jm9811007
- Primary Citation of Related Structures:
1BM2, 1BMB - PubMed Abstract:
Following earlier work on cystine-bridged peptides, cyclic phosphopeptides containing nonreducible mimics of cystine were synthesized that show high affinity and specificity toward the Src homology (SH2) domain of the growth factor receptor-binding protein (Grb2). Replacement of the cystine in the cyclic heptapeptide cyclo(CYVNVPC) by D-alpha-acetylthialysine or D-alpha-lysine gave cyclo(YVNVP(D-alpha-acetyl-thiaK)) (22) and cyclo(YVNVP(D-alpha-acetyl-K)) (30), which showed improved binding 10-fold relative to that of the control peptide KPFYVNVEF (1). NMR spectroscopy and molecular modeling experiments indicate that a beta-turn conformation centered around YVNV is essential for high-affinity binding. X-ray structure analyses show that the linear peptide 1 and the cyclic compound 21 adopt a similar binding mode with a beta-turn conformation. Our data confirm the unique structural requirements of the ligand binding site of the SH2 domain of Grb2. Moreover, the potency of our cyclic lactams can be explained by the stabilization of the beta-turn conformation by three intramolecular hydrogen bonds (one mediated by an H2O molecule). These stable and easily accessible cyclic peptides can serve as templates for the evaluation of phosphotyrosine surrogates and further chemical elaboration.
- Novartis Forschungsinstitut, Brunnerstrasse 59, Vienna A-1235, Austria. peter.ettmayer@pharma.novartis.com
Organizational Affiliation: