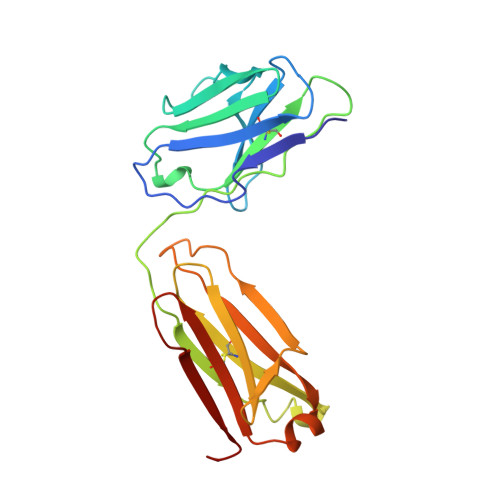
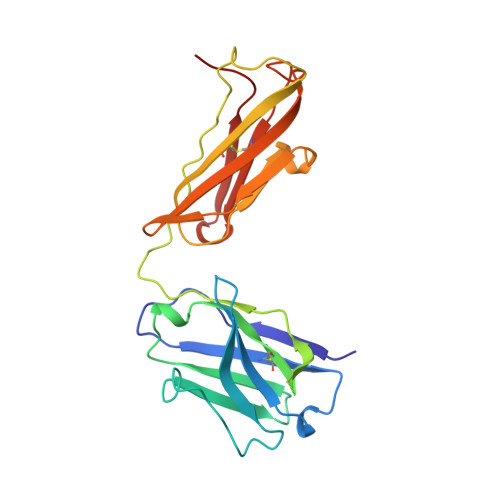
Conformational change in an anti-integrin antibody: structure of OPG2 Fab bound to a beta 3 peptide.
Kodandapani, R., Veerapandian, L., Ni, C.Z., Chiou, C.K., Whittal, R.M., Kunicki, T.J., Ely, K.R.(1998) Biochem Biophys Res Commun 251: 61-66
- PubMed: 9790907
- DOI: https://doi.org/10.1006/bbrc.1998.9380
- Primary Citation of Related Structures:
1BM3 - PubMed Abstract:
Antibodies are important tools to explore receptor-ligand interactions. The anti-integrin antibody OPG2 binds in an RGD-related manner to the alphaIIb beta3 integrin as a molecular mimic of fibrinogen. The Fab fragment from OPG2 was cocrystallized with a peptide from the beta3 subunit of the integrin representing a site that binds RGD. The crystal structure of the complex was determined at 2.2-A resolution and compared with the unbound Fab. On binding the integrin peptide there were conformational changes in CDR3 of the heavy chain. Also, a significant shift across the intermolecular interface between the CH1-CL domains was observed so that the angle of rotation relating the two domains was reduced by 15 degrees. This unusual conformational adjustment represents the first example of ligand-induced conformational changes in the carboxyl domains of a Fab fragment.
- Cancer Research Center, The Burnham Institute, La Jolla, California, 92037, USA.
Organizational Affiliation: