Three-dimensional solution structure of the B domain of staphylococcal protein A: comparisons of the solution and crystal structures.
Gouda, H., Torigoe, H., Saito, A., Sato, M., Arata, Y., Shimada, I.(1992) Biochemistry 31: 9665-9672
- PubMed: 1390743
- DOI: https://doi.org/10.1021/bi00155a020
- Primary Citation of Related Structures:
1BDC, 1BDD - PubMed Abstract:
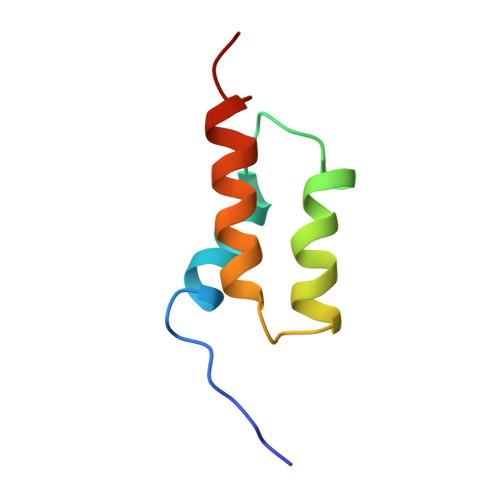
The three-dimensional solution structure of the recombinant B domain (FB) of staphylococcal protein A, which specifically binds to the Fc portion of immunoglobulin G, was determined by NMR spectroscopy and hybrid distance geometry-dynamical simulated annealing calculations. On the basis of 692 experimental constraints including 587 distance constraints obtained from the nuclear Overhauser effect (NOE), 57 torsion angle (phi, chi 1) constraints, and 48 constraints associated with 24 hydrogen bonds, a total of 10 converged structures of FB were obtained. The atomic root mean square difference among the 10 converged structures is 0.52 +/- 0.10 A for the backbone atoms and 0.98 +/- 0.08 A for all heavy atoms (excluding the N-terminal segment from Thr1 to Glu9 and the C-terminal segment from Gln56 to Ala60, which are partially disordered). FB is composed of a bundle of three alpha-helices, i.e., helix I (Gln10-His19), helix II (Glu25-Asp37), and helix III (Ser42-Ala55). Helix II and helix III are antiparallel to each other, whereas the long axis of helix I is tilted at an angle of about 30 degrees with respect to those of helix II and helix III. Most of the hydrophobic residues of FB are buried in the interior of the bundle of the three helices. It is suggested that the buried hydrophobic residues form a hydrophobic core, contributing to the stability of FB.(ABSTRACT TRUNCATED AT 250 WORDS)
- Faculty of Pharmaceutical Sciences, University of Tokyo, Japan.
Organizational Affiliation: