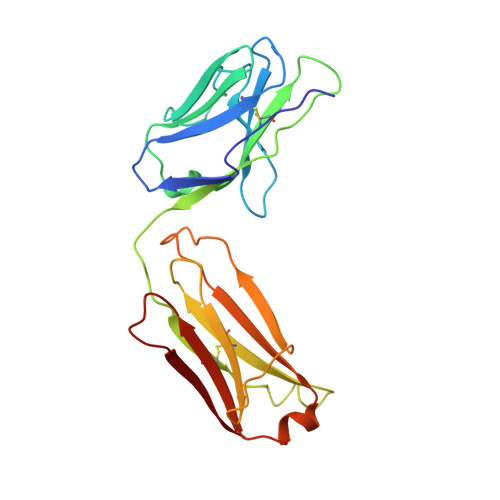
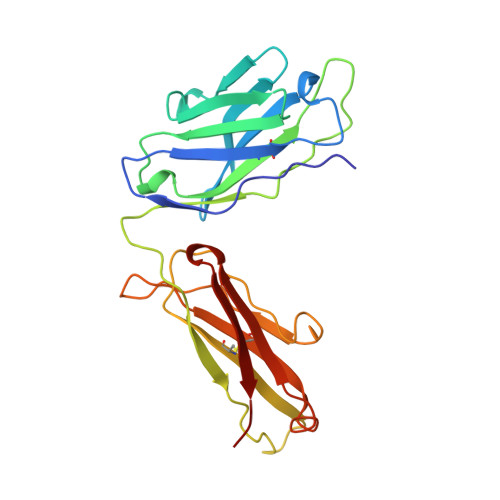
The interplay between binding energy and catalysis in the evolution of a catalytic antibody.
Ulrich, H.D., Mundorff, E., Santarsiero, B.D., Driggers, E.M., Stevens, R.C., Schultz, P.G.(1997) Nature 389: 271-275
- PubMed: 9305839
- DOI: https://doi.org/10.1038/38470
- Primary Citation of Related Structures:
1AXS - PubMed Abstract:
Antibody catalysis provides an opportunity to examine the evolution of binding energy and its relation to catalytic function in a system that has many parallels with natural enzymes. Here we report such a study involving an antibody AZ-28 that catalyses an oxy-Cope rearrangement, a pericyclic reaction that belongs to a well studied and widely used class of reactions in organic chemistry. Immunization with transition state analogue 1 results in a germline-encoded antibody that catalyses the rearrangement of hexadiene 2 to aldehyde 3 with a rate approaching that of a related pericyclic reaction catalysed by the enzyme chorismate mutase. Affinity maturation gives antibody AZ-28, which has six amino acid substitutions, one of which results in a decrease in catalytic rate. To understand the relationship between binding and catalytic rate in this system we characterized a series of active-site mutants and determined the three-dimensional crystal structure of the complex of AZ-28 with the transition state analogue. This analysis indicates that the activation energy depends on a complex balance of several stereoelectronic effects which are controlled by an extensive network of binding interactions in the active site. Thus in this instance the combinatorial diversity of the immune system provided both an efficient catalyst for a reaction where no enzyme is known, as well as an opportunity to explore the mechanisms and evolution of biological catalysis.
- Howard Hughes Medical Institute, University of California, Berkeley 94720, USA.
Organizational Affiliation: