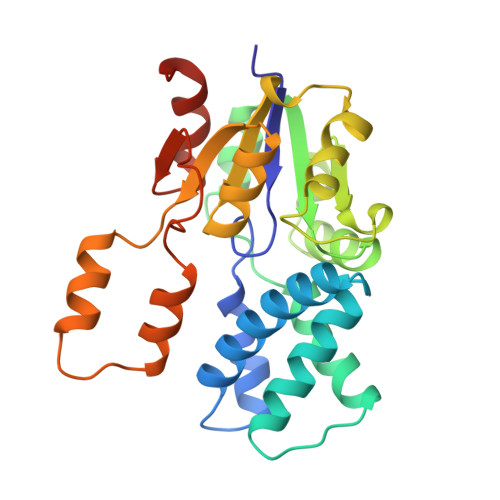
Three-dimensional structure of L-2-haloacid dehalogenase from Xanthobacter autotrophicus GJ10 complexed with the substrate-analogue formate.
Ridder, I.S., Rozeboom, H.J., Kalk, K.H., Janssen, D.B., Dijkstra, B.W.(1997) J Biological Chem 272: 33015-33022
- PubMed: 9407083
- DOI: https://doi.org/10.1074/jbc.272.52.33015
- Primary Citation of Related Structures:
1AQ6 - PubMed Abstract:
The L-2-haloacid dehalogenase from the 1,2-dichloroethane degrading bacterium Xanthobacter autotrophicus GJ10 catalyzes the hydrolytic dehalogenation of small L-2-haloalkanoic acids to yield the corresponding D-2-hydroxyalkanoic acids. Its crystal structure was solved by the method of multiple isomorphous replacement with incorporation of anomalous scattering information and solvent flattening, and was refined at 1.95-A resolution to an R factor of 21.3%. The three-dimensional structure is similar to that of the homologous L-2-haloacid dehalogenase from Pseudomonas sp. YL (1), but the X. autotrophicus enzyme has an extra dimerization domain, an active site cavity that is completely shielded from the solvent, and a different orientation of several catalytically important amino acid residues. Moreover, under the conditions used, a formate ion is bound in the active site. The position of this substrate-analogue provides valuable information on the reaction mechanism and explains the limited substrate specificity of the Xanthobacter L-2-haloacid dehalogenase.
- Laboratory of Biophysical Chemistry, Department of Chemistry, University of Groningen, Nijenborgh 4, 9747 AG Groningen, The Netherlands.
Organizational Affiliation: