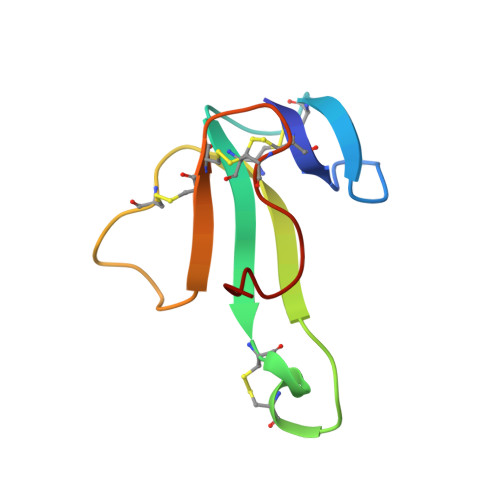
NMR solution structure of an alpha-bungarotoxin/nicotinic receptor peptide complex.
Basus, V.J., Song, G., Hawrot, E.(1993) Biochemistry 32: 12290-12298
- PubMed: 8241115
- DOI: https://doi.org/10.1021/bi00097a004
- Primary Citation of Related Structures:
1ABT - PubMed Abstract:
We report the two-dimensional nuclear magnetic resonance (NMR) characterization of the stoichiometric complex formed between the snake venom-derived long alpha-neurotoxin, alpha-bungarotoxin (BGTX), and a synthetic dodecapeptide (alpha 185-196) corresponding to a functionally important region on the alpha-subunit of the nicotinic acetylcholine receptor (nAChR) obtained from Torpedo californica electric organ tissue. BGTX has been widely used as the classic nicotinic competitive antagonist for the skeletal muscle type of nAChR which is found in the avian, amphibian, and mammalian neuromuscular junction. The receptor dodecapeptide (alpha 185-196) binds BGTX with micromolar affinity and has been shown to represent the major determinant of BGTX binding to the isolated alpha-subunit. Previous studies involving covalent modification of the native nAChR from Torpedo membranes with a variety of affinity reagents indicate that several residues contained within the dodecapeptide sequence (namely, Tyr-190, Cys-192, and Cys-193) apparently contribute directly to the formation of the cholinergic ligand binding site. The NMR-derived solution structure of the BGTX/receptor peptide complex defines a relatively extended conformation for a major segment of the "bound" dodecapeptide. These structural studies also reveal a previously unpredicted receptor binding cleft within BGTX and suggest that BGTX undergoes a conformational change upon peptide binding. If, as we hypothesize, the identified intermolecular contacts in the BGTX/receptor peptide complex describe a portion of the contact zone between BGTX and native receptor, then the structural data would suggest that alpha-subunit residues 186-190 are on the extracellular surface of the receptor.
- Department of Pharmaceutical Chemistry, University of California, San Francisco 94143.
Organizational Affiliation: