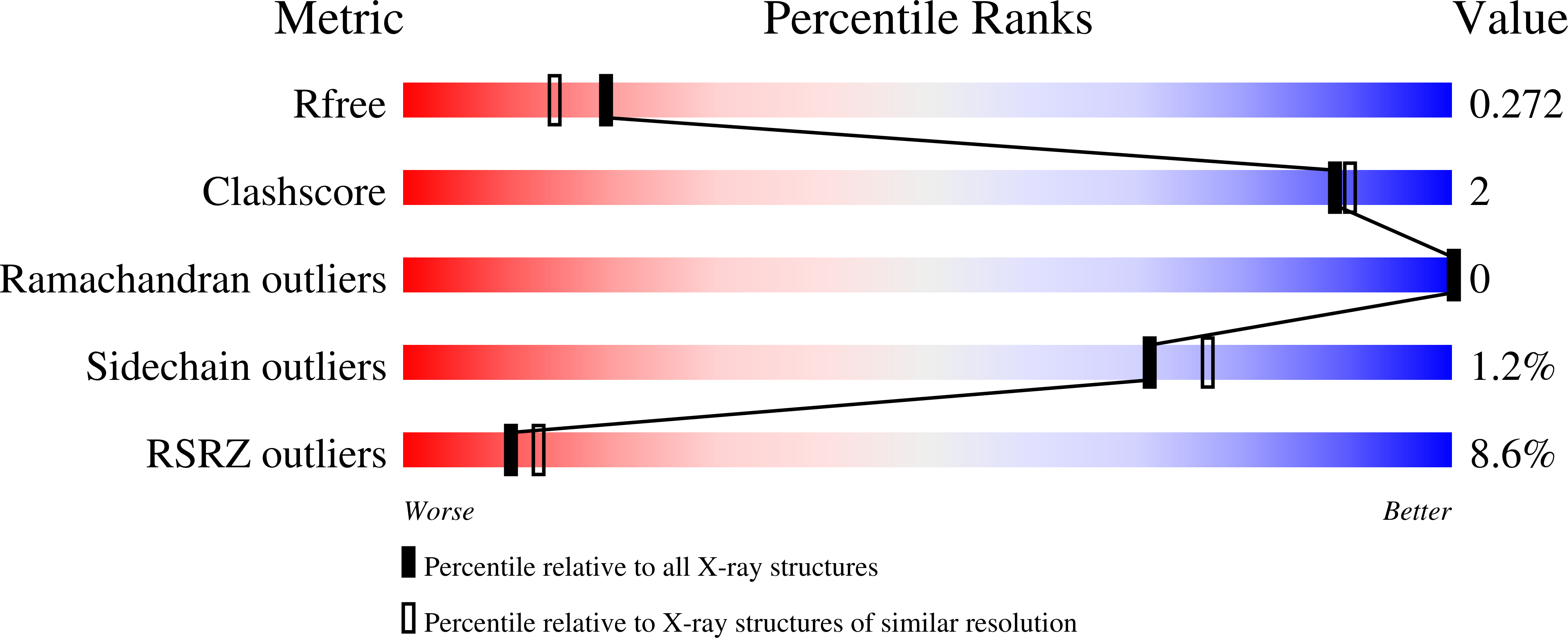
Discovery of AZD4831, a Mechanism-Based Irreversible Inhibitor of Myeloperoxidase, As a Potential Treatment for Heart Failure with Preserved Ejection Fraction.
Inghardt, T., Antonsson, T., Ericsson, C., Hovdal, D., Johannesson, P., Johansson, C., Jurva, U., Kajanus, J., Kull, B., Michaelsson, E., Pettersen, A., Sjogren, T., Sorensen, H., Westerlund, K., Lindstedt, E.L.(2022) J Med Chem 65: 11485-11496
- PubMed: 36005476
- DOI: https://doi.org/10.1021/acs.jmedchem.1c02141
- Primary Citation of Related Structures:
7NI1, 7NI3 - PubMed Abstract:
Myeloperoxidase is a promising therapeutic target for treatment of patients suffering from heart failure with preserved ejection fraction (HFpEF). We aimed to discover a covalent myeloperoxidase inhibitor with high selectivity for myeloperoxidase over thyroid peroxidase, limited penetration of the blood-brain barrier, and pharmacokinetics suitable for once-daily oral administration at low dose. Structure-activity relationship, biophysical, and structural studies led to prioritization of four compounds for in-depth safety and pharmacokinetic studies in animal models. One compound (AZD4831) progressed to clinical studies on grounds of high potency (IC 50 , 1.5 nM in vitro ) and selectivity (>450-fold vs thyroid peroxidase in vitro ), the mechanism of irreversible inhibition, and the safety profile. Following phase 1 studies in healthy volunteers and a phase 2a study in patients with HFpEF, a phase 2 b /3 efficacy study of AZD4831 in patients with HFpEF started in 2021.
Organizational Affiliation:
Research and Early Development, Cardiovascular, Renal and Metabolism, BioPharmaceuticals R&D, AstraZeneca, 431 50 Mölndal, Gothenburg, Sweden.