Insights into the Neutralization and DNA Binding of Toxin-Antitoxin System ParE SO-CopA SO by Structure-Function Studies.
Zhou, J., Du, X.J., Liu, Y., Gao, Z.Q., Geng, Z., Dong, Y.H., Zhang, H.(2021) Microorganisms 9
- PubMed: 34946107
- DOI: https://doi.org/10.3390/microorganisms9122506
- Primary Citation of Related Structures:
7ETR - PubMed Abstract:
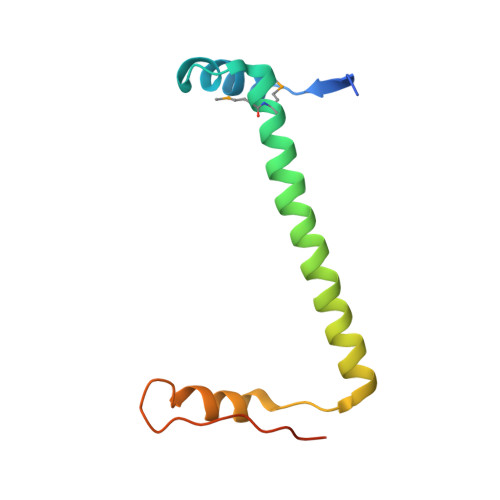
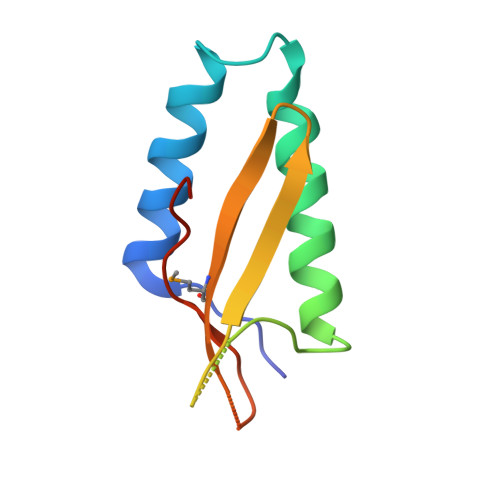
ParE SO -CopA SO is a new type II toxin-antitoxin (TA) system in prophage CP4So that plays an essential role in circular CP4So maintenance after the excision in Shewanella oneidensis . The toxin ParE SO severely inhibits cell growth, while CopA SO functions as an antitoxin to neutralize ParE SO toxicity through direct interactions. However, the molecular mechanism of the neutralization and autoregulation of the TA operon transcription remains elusive. In this study, we determined the crystal structure of a ParE SO -CopA SO complex that adopted an open V-shaped heterotetramer with the organization of ParE SO -(CopA SO ) 2 -ParE SO . The structure showed that upon ParE SO binding, the intrinsically disordered C-terminal domain of CopA SO was induced to fold into a partially ordered conformation that bound into a positively charged and hydrophobic groove of ParE SO . Thermodynamics analysis showed the DNA-binding affinity of CopA SO was remarkably higher than that of the purified TA complex, accompanied by the enthalpy change reversion from an exothermic reaction to an endothermic reaction. These results suggested ParE SO acts as a de-repressor of the TA operon transcription at the toxin:antitoxin level of 1:1. Site-directed mutagenesis of ParE SO identified His91 as the essential residue for its toxicity by cell toxicity assays. Our structure-function studies therefore elucidated the transcriptional regulation mechanism of the ParE SO -CopA SO pair, and may help to understand the regulation of CP4So maintenance in S. oneidensis .
Organizational Affiliation:
Institute of Health Sciences and School of Life Science, Anhui University, Hefei 230601, China.