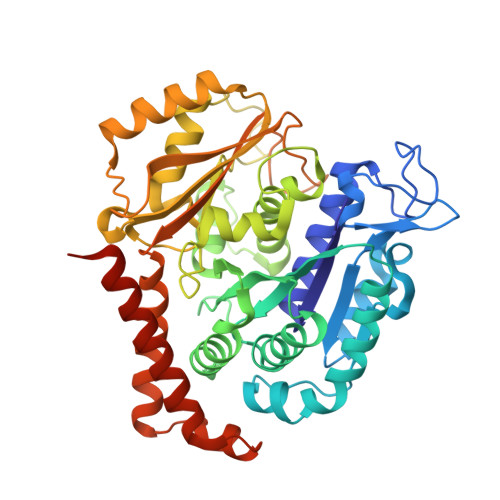
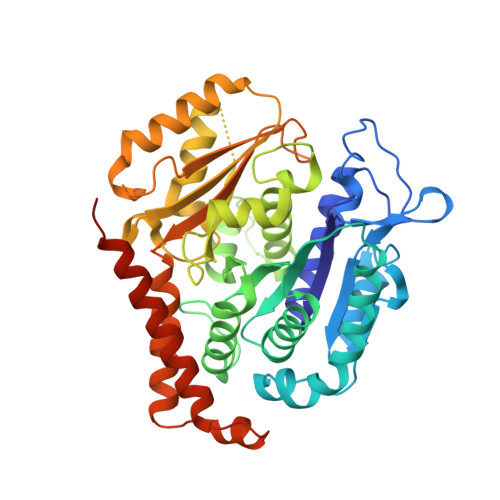
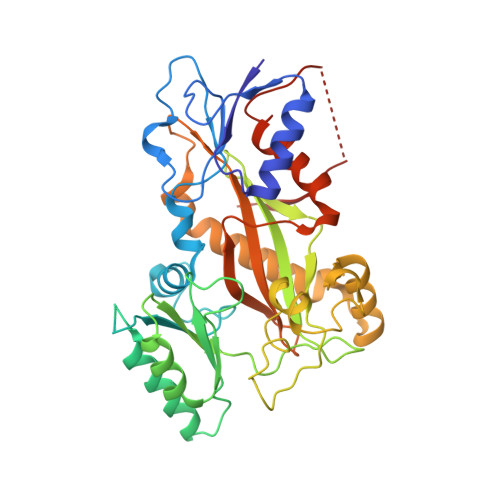
Cevipabulin-tubulin complex reveals a novel agent binding site on alpha-tubulin with tubulin degradation effect.
Yang, J., Yu, Y., Li, Y., Yan, W., Ye, H., Niu, L., Tang, M., Wang, Z., Yang, Z., Pei, H., Wei, H., Zhao, M., Wen, J., Yang, L., Ouyang, L., Wei, Y., Chen, Q., Li, W., Chen, L.(2021) Sci Adv 7
- PubMed: 34138737
- DOI: https://doi.org/10.1126/sciadv.abg4168
- Primary Citation of Related Structures:
7CLD, 7DP8 - PubMed Abstract:
Microtubules, composed of αβ-tubulin heterodimers, have remained popular anticancer targets for decades. Six known binding sites on tubulin dimers have been identified thus far, with five sites on β-tubulin and only one site on α-tubulin, hinting that compounds binding to α-tubulin are less well characterized. Cevipabulin, a microtubule-active antitumor clinical candidate, is widely accepted as a microtubule-stabilizing agent by binding to the vinblastine site. Our x-ray crystallography study reveals that, in addition to binding to the vinblastine site, cevipabulin also binds to a new site on α-tubulin. We find that cevipabulin at this site pushes the αT5 loop outward, making the nonexchangeable GTP exchangeable, which reduces the stability of tubulin, leading to its destabilization and degradation. Our results confirm the existence of a new agent binding site on α-tubulin and shed light on the development of tubulin degraders as a new generation of antimicrotubule drugs targeting this novel site.
Organizational Affiliation:
Laboratory of Natural and Targeted Small Molecule Drugs, State Key Laboratory of Biotherapy and Cancer Center, National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University and Collaborative Innovation Center of Biotherapy, Chengdu 610041, China. chenlijuan125@163.com weimi003@yahoo.com yjh1988@scu.edu.cn.